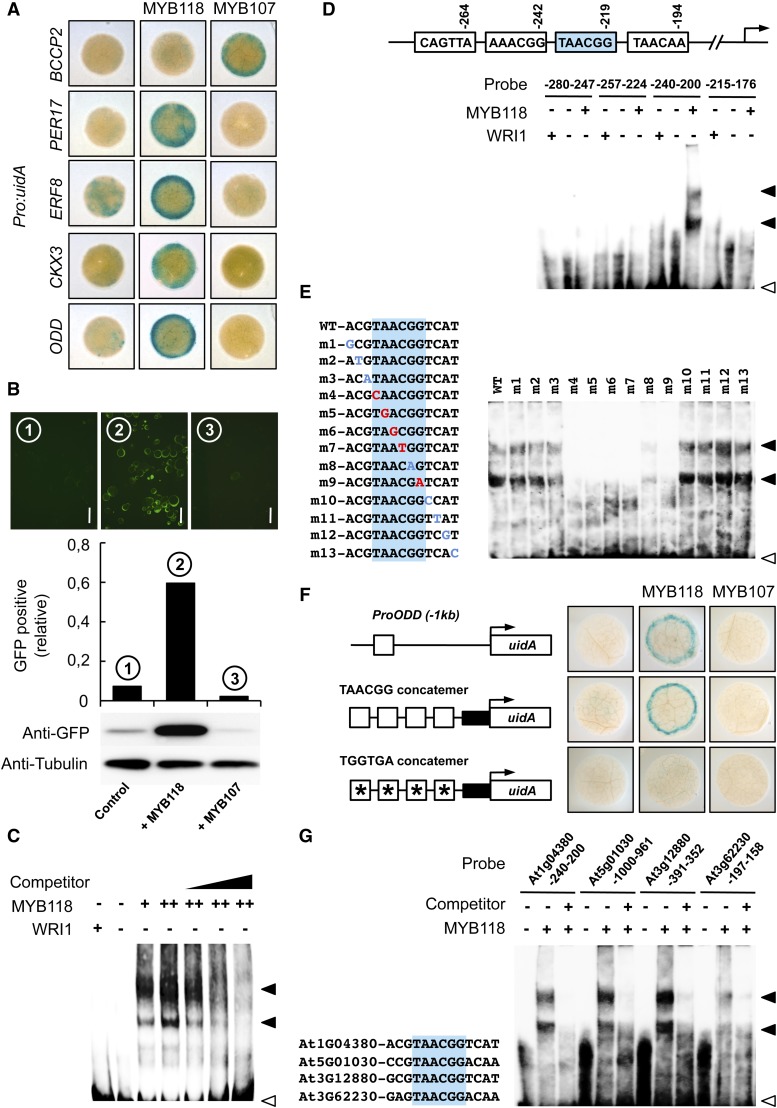
Figure 6.
Transcriptional Activation of MYB118 Targets.
(A) Transactivation assay in leaves of N. benthamiana. Pro:uidA reporter constructs alone or in combination with a vector allowing the expression of MYB118 or MYB107 (negative control) were coinfiltrated in young leaves of N. benthamiana with a vector allowing the expression of the p19 protein of tomato bushy stunt virus (TBSV) that prevents the onset of posttranscriptional gene silencing (Voinnet et al., 2003). Leaf discs were assayed for GUS activity 3 d after infiltration. Tissues were incubated 5 h in a buffer containing 0.2 mM each of potassium ferrocyanide and potassium ferricyanide. Representative discs (diameter = 0.8 cm) are presented.
(B) Transactivation assay in protoplasts of Arabidopsis. Protoplasts were transformed with the ProODD:GFP reporter construct alone or in combination with a vector allowing the expression of MYB118 or MYB107 (negative control). Whole mounts of protoplasts were observed 12 h after transformation and the relative proportion of GFP-expressing protoplasts was calculated (between 800 and 1000 protoplasts were scored). Immunoblots of a batch of 0.5 mL transformed protoplasts for each combination of constructs assayed and using primary antibodies raised against GFP (anti-GFP serum) or α-tubulin (anti-Tubulin serum) are presented. Bars = 50 µm.
(C) Binding of MYB118 to the proximal upstream region of ODD. EMSA of a probe covering a region from −280 to −191 bp upstream from the ATG codon of ODD with increasing amounts of MYB118 (+ = 0.4 µg and ++ = 0.6 µg). WRI1 was used as a negative control. Competition of MYB118 binding was performed in the presence of 1-, 5-, and 10-fold amounts of the unlabeled ProODD(-280 to -191 bp) fragment. Positions of free probe (open arrowhead) and the shifted bands (closed arrowheads) are indicated.
(D) Identification of the binding site of MYB118 in the proximal upstream region of ODD. A schematic representation of the 280-bp ODD promoter showing the position of putative MYB-core cis-regulatory elements is presented. Indicated positions correspond to the distance of the end of these elements relative to the ATG start codon in base pairs. The EMSA of four partially overlapping sequences containing one of these elements each is shown below the representation.
(E) Binding site sequence specificity of MYB118. The EMSA of MYB118 binding to ProODD(-240 to -201 bp) probes containing single mutations near the MYB-core element (shaded in blue) is presented. Mutations that affected MYB118 binding are indicated in red, while mutations that did not affect MYB118 binding are indicated in blue.
(F) Transactivation by MYB118 of a promoter sequence made of a concatemer of TAACGG elements separated by 10 nucleotides. Schematic representations of the reporter constructs used are presented. Open boxes indicate TAACGG elements, and asterisks indicate that the MYB-core element was modified as indicated. Closed boxes represent the 35S cauliflower mosaic virus minimal promoter. Reporter constructs alone or in combination with a vector allowing the expression of MYB118 or MYB107 (negative control) were coinfiltrated in young leaves of N. benthamiana with a vector allowing the expression of the p19 protein of tomato bushy stunt virus (TBSV). Leaf discs were assayed for GUS activity 3 d after infiltration. Tissues were incubated 2 h in a buffer containing 2 mM each of potassium ferrocyanide and potassium ferricyanide. Representative discs (diameter = 0.5 cm) are presented.
(G) Binding of MYB118 to the proximal upstream regions of genes downregulated in myb118-1 seeds.