Paternal hypomethylation can bypass the interploidy hybridization barrier by alleviating the requirement for the epigenetic Polycomb Repressive Complex 2 (PRC2) in the endosperm. The hypomethylated pollen genome causes de novo CHG methylation directed to FIS-PRC2 target genes, suggesting that different epigenetic modifications can functionally substitute for each other.
Abstract
Plants of different ploidy levels are separated by a strong postzygotic hybridization barrier that is established in the endosperm. Deregulated parent-of-origin specific genes cause the response to interploidy hybridizations, revealing an epigenetic basis of this phenomenon. In this study, we present evidence that paternal hypomethylation can bypass the interploidy hybridization barrier by alleviating the requirement for the Polycomb Repressive Complex 2 (PRC2) in the endosperm. PRC2 epigenetically regulates gene expression by applying methylation marks on histone H3. Bypass of the barrier is mediated by suppressed expression of imprinted genes. We show that the hypomethylated pollen genome causes de novo CHG methylation directed to FIS-PRC2 target genes, suggesting that different epigenetic modifications can functionally substitute for each other. Our work presents a method for the generation of viable triploids, providing an impressive example of the potential of epigenome manipulations for plant breeding.
INTRODUCTION
Postzygotic reproductive isolation in response to hybridization between individuals of the same plant species at different ploidy levels (interploidy hybridizations) forms a major path for sympatric speciation in plants (Ramsey and Schemske, 1998; Otto and Whitton, 2000). This phenomenon has been termed triploid block and operates in the endosperm, a terminal tissue that surrounds and nourishes the embryo (Brink and Cooper, 1947; Marks, 1966; Ramsey and Schemske, 1998). The endosperm of most angiosperms is triploid (3n) and is derived from the fertilization of the homodiploid central cell by one haploid sperm cell, whereas the diploid (2n) embryo is the result of the fertilization of the haploid egg cell by the second haploid sperm cell (Drews and Yadegari, 2002; Li and Berger, 2012). Given the nature of this double fertilization, the embryo contains one maternal and one paternal (1m:1p) genome copy, whereas the endosperm consists of two maternal and one paternal copies (2m:1p). In interploidy hybridizations, the parental genome contributions are altered, adding either one additional maternal or paternal copy to the embryo and endosperm, thereby creating either maternal or paternal excess hybridizations depending on whether the ovule donor or the pollen donor has a higher ploidy level. The parental genome dosage affects endosperm cellularization, a process essential for the formation of viable seeds (Scott et al., 1998; Hehenberger et al., 2012; Kradolfer et al., 2013a).
Parental dosage sensitivity of the endosperm led to the hypothesis that imprinted genes are causal for the response to interploidy hybridizations (Haig and Westoby, 1989; Birchler, 1993; Adams et al., 2000; Gutierrez-Marcos et al., 2003; Kinoshita, 2007). Imprinted genes are specifically or preferentially expressed from only one parental allele, generating the basis for dosage-sensitive behavior upon changes of parental genome contributions. Imprinted gene expression depends on the activity of the FERTILIZATION INDEPENDENT SEED (FIS) Polycomb Repressive Complex 2 (PRC2) and the specific establishment and erasure of DNA methylation marks (Pignatta and Gehring, 2012). Target gene repression by PRC2 is mediated by trimethylation of lysine 27 of histone H3 (H3K27m3) (Simon and Kingston, 2013). Mutants in FIS-PRC2 subunits phenotypically mimic triploid seeds, suggesting that deregulation of FIS-PRC2 target genes are causally responsible for triploid seed failure (Erilova et al., 2009; Tiwari et al., 2010). Supporting this view, mutations in the paternally expressed imprinted FIS-PRC2 target gene ADMETOS (ADM) can suppress triploid seed abortion (Kradolfer et al., 2013b).
DNA methylation in plants can occur in different sequence contexts featuring symmetric CG and CHG methylation as well as asymmetric CHH methylation (where H can be C, T, or A) (Law and Jacobsen, 2010). DNA methylation in a CG context is maintained by the methyltransferase MET1, whereas CHG methylation is maintained by the chromomethylase CMT3 and requires activity of the H3K9 dimethyltransferase KRYPTONITE (Lindroth et al., 2001; Jackson et al., 2002). Establishment and maintenance of CHH methylation requires the de novo methyltransferase DOMAINS REARRANGED2 (DRM2) and the chromomethylase CMT2. While recruitment of DRM2 depends on 24-nucleotide small interfering RNAs, CMT2 is recruited via dimethylation of histone H3 lysine 9 (H3K9m2) (Law and Jacobsen, 2010; Zemach et al., 2013; Stroud et al., 2014).
Parent-of-origin allele-specific methylation differences that underpin imprinted gene regulation are a consequence of the central cell specific DNA demethylation activity of the DEMETER DNA glycosylase that excises 5-methylcytosine. As a consequence, the maternal genome in the endosperm is characterized by genome-wide hypomethylation that particularly affects short transposable elements (TEs) and repeats (Gehring et al., 2009; Hsieh et al., 2009). Many differentially methylated TEs and repeats reside near genes and cause imprinted expression (Gehring et al., 2011; Hsieh et al., 2011; Wolff et al., 2011).
Hypomethylated pollen derived from the met1 mutant causes precocious endosperm cellularization and decreased seed size (Xiao et al., 2006), which may be a consequence of suppressed activity of paternally expressed imprinted genes (Hsieh et al., 2011). Deregulated expression of the paternally expressed imprinted gene ADM was shown to be causally responsible for triploid seed abortion (Kradolfer et al., 2013b); therefore, we raised and tested the hypothesis that paternal CG hypomethylation bypasses the interploidy hybridization barrier by suppressing deregulated imprinted genes. Hypomethylated pollen can also bypass the maternal FIS-PRC2 requirement (Luo et al., 2000; Vinkenoog et al., 2000), raising the hypothesis that triploid seed rescue may be independent of a functional FIS-PRC2. Here, we report that met1 pollen can suppress triploid seed abortion and bypass the requirement of FIS-PRC2, allowing formation of viable triploid seeds at high frequency. We show that the bypass of FIS-PRC2 by met1 correlates with de novo CHG methylation directed to FIS-PRC2 target genes, suggesting that different epigenetic modifications can functionally substitute for each other. Our work has the potential to make a substantial impact on plant breeding, as it provides a method for the generation of viable triploids, which otherwise require laborious embryo rescue methods, providing an impressive example of the potential of epigenome manipulations for plant breeding.
RESULTS
met1-Induced Paternal Genome Hypomethylation Suppresses Triploid Seed Abortion
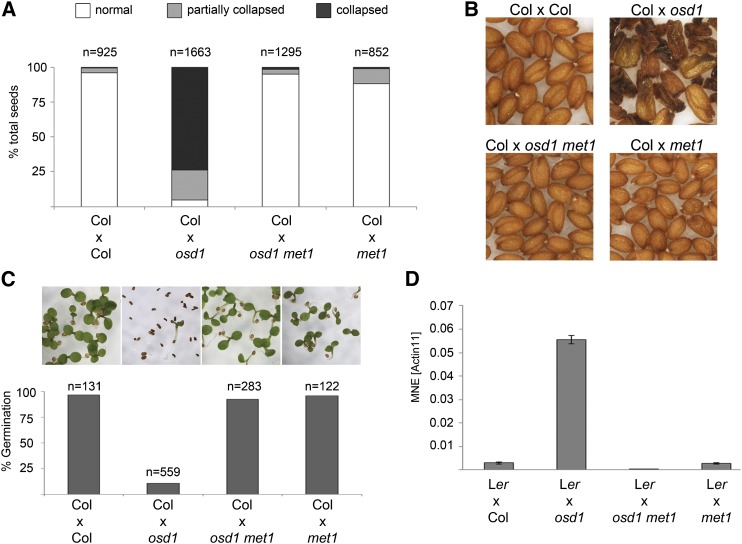
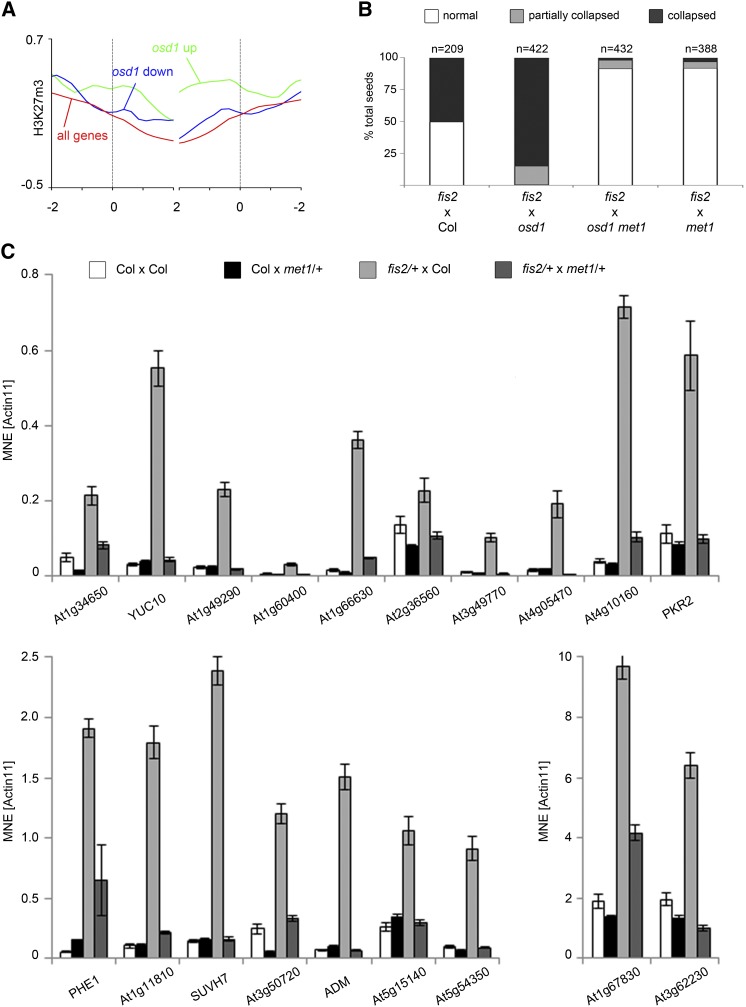
To test whether paternal CG hypomethylation may bypass the triploid block, we generated double mutants of met1 and omission of second division1 (osd1) and pollinated wild-type Columbia (Col) plants with met1 osd1 double mutant pollen. The osd1 mutant forms 100% unreduced (2n) male gametes that when crossed to wild-type plants lead to 100% triploid seeds that abort at high frequency (d’Erfurth et al., 2009; Kradolfer et al., 2013b). Indeed, we observed very strong suppression of triploid seed abortion using hypomethylated unreduced pollen; while pollination with osd1-derived 2n pollen gave rise to 95% of seeds that were either partially or totally collapsed, osd1 met1-derived 2n pollen suppressed triploid seed abortion to 1.5% (Figure 1A). Rescued triploid seeds had a wild-type-like shape and size and germinated at high frequency (Figures 1B and 1C; Supplemental Figure 1A). Expression of ADM was suppressed in seeds derived from pollination with 2n met1 pollen, correlating with normalized triploid seed development (Figure 1D).
Figure 1.
Bypass of Seed Abortion by Hypomethylation of the Paternal Genome.
(A) Percentage of collapsed and partially collapsed seeds in interploidy hybridizations. n, total number of seeds tested.
(B) Phenotype of mature seeds. Genotypes are indicated above.
(C) Percentage of germinating seeds in hybridizations with different paternal genotypes. Pictures correspond to columns in the graph below; n, total number of seeds tested.
(D) Expression of ADM in seeds 6 DAP. MNE, mean normalized expression. ACTIN11 was used as reference. Error bars indicate se.
[See online article for color version of this figure.]
To test whether suppression of triploid seed abortion by met1 occurs independently of the osd1 mutation, we generated tetraploid (4X) met1 mutant plants using colchicine treatment. Similarly, pollination of wild-type plants with pollen from three independent 4X met1 mutants caused strong suppression of triploid block (Supplemental Figure 2), revealing that the observed effect of met1-induced hypomethylated 2n pollen on triploid seed rescue is independent of the osd1 mutation.
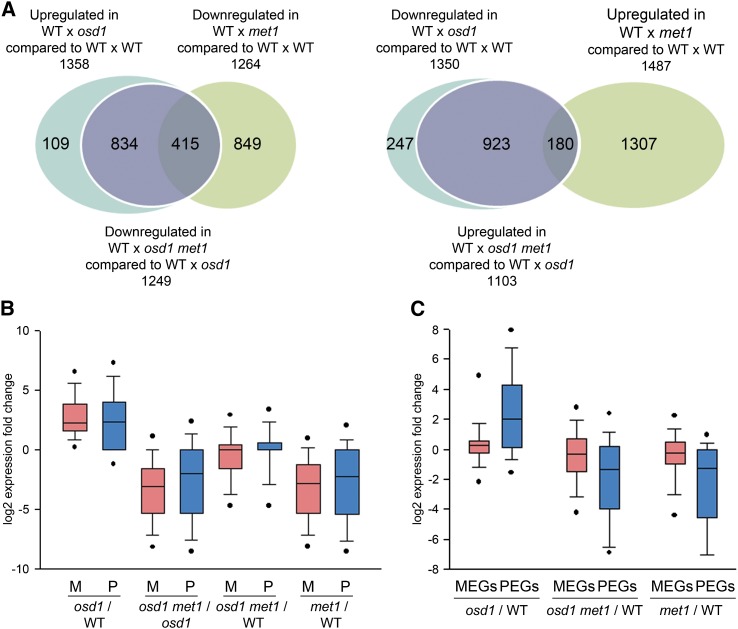
To test whether rescue of seed development was reflected by a genome-wide normalization of gene expression, we generated genome-wide expression data of seeds derived from crosses Ler × Col, Ler × osd1, Ler × osd1 met1, and Ler × met1 that were harvested at 6 d after pollination (DAP). The accession Landsberg erecta (Ler) was used as the maternal parent to enable assignment of parent-specific expression changes. The vast majority of genes that were deregulated in response to interploidy hybridization were normalized in seeds derived from osd1 met1 pollinations (Figure 2A; Supplemental Data Set 1). The suppressive effect of met1 was not restricted to triploid seeds, since transcript levels of many genes were reduced in seeds derived from pollination of wild-type plants with met1 pollen. About 30% of those repressed genes affected by met1 pollination overlapped with genes that were deregulated in triploid seeds (hypergeometric test P = 1.4E-268; Figure 2A, left panel), revealing that paternal hypomethylation suppresses dosage-sensitive genes.
Figure 2.
Deregulated Genes in Interploidy Hybridizations Are Normally Expressed in Hypomethylated Paternal Excess Hybridizations.
(A) Left panel: Venn diagram showing overlap of genes being upregulated in seeds derived from wild type × osd1 crosses (signal log ratio [SLR] > 1, P < 0.05) with genes that are downregulated in wild type × met1 (SLR < −1, P < 0.05) and genes that are downregulated in wild type × osd1 met1 compared with wild type × osd1 (SLR < −1, P < 0.05). Right panel: Venn diagram showing overlap of genes being downregulated in seeds derived from wild type × osd1 crosses with genes that are upregulated in wild type × met1 and genes that are upregulated in wild type × osd1 met1 compared with wild type × osd1 (SLR > 1, P < 0.05).
(B) Expression fold change of maternal (M; red bars) and paternal (P; blue bars) alleles of genes being upregulated in crosses wild type × osd1 versus wild type × wild type; genes being downregulated in crosses wild type × osd1 met1 versus wild type × osd1; genes being downregulated in crosses wild type × osd1 met1 versus wild type × wild type; and genes being downregulated in crosses wild type × met1 versus wild type × wild type. SLRs and P values of crosses are as indicated in (A).
(C) Expression of MEGs (red bars) and PEGs (blue bars) in crosses of wild type × osd1 versus wild type × wild type; wild type × osd1 met1 versus wild type × wild type; and wild type × met1 versus wild type × wild type.
[See online article for color version of this figure.]
We specifically analyzed genes that were previously shown to be deregulated in triploid seeds and in interspecific hybrid seeds (Tiwari et al., 2010; Burkart-Waco et al., 2013; Kradolfer et al., 2013b) (Supplemental Figure 3). Expression of all analyzed type I MADS box transcription factor genes, as well as HAIKU genes (IKU1 and IKU2) and TRANSPARENT TESTA GLABRA2, was suppressed by osd1 met1 pollen, consistent with a normalization of triploid seed development. Importantly, all genes were also suppressed in diploid seeds derived from met1 pollen, suggesting that the same mechanism underlies met1-mediated gene repression in diploid and triploid seeds. Expression of FIS-PRC2 subunit encoding genes MEDEA (MEA) and FIS2 was suppressed by met1 and osd1 met1 pollen, suggesting that normalization of triploid seed development by osd1 met1 pollen is not a consequence of normalized MEA and FIS2 expression. Mutants in NONEXPRESSOR OF PR GENES (NPR1) and SALICYLIC ACID INDUCTION-DEFICIENT2 (SID2) were previously shown to suppress hybrid seed abortion (Burkart-Waco et al., 2013). However, expression of NPR1 was further increased in osd1 met1 and met1 derived seeds compared with osd1 and wild-type pollinated controls, while expression of SID2 was largely unchanged in the corresponding crosses, suggesting that both genes are unlikely to play a causal role in restoring triploid seed rescue.
We further tested whether increased paternal genome dosage caused preferentially increased expression of one of the parental alleles. Expression of both alleles was increased in triploid seeds and suppressed to wild-type levels by hypomethylated 2n pollen (Figure 2B). There was the tendency of the maternal alleles of seeds pollinated with hypomethylated pollen being stronger affected compared with the paternal alleles, which was clearly detectable in triploid and diploid seeds. These findings suggest that the suppressive effect of met1 was established after fertilization. We also tested for preferential deregulation of maternally or paternally expressed imprinted genes (MEGs or PEGs, respectively) in response to increased paternal ploidy and hypomethylation. While MEGs were only slightly affected in response to increased paternal ploidy and hypomethylation (Figure 2C), PEGs were strongly upregulated in response to increased ploidy and repressed by hypomethylated 2n and 1n pollen (Figure 2C), suggesting that different regulatory mechanisms act on MEGs and PEGs in response to interploidy hybridizations.
Increased Sperm Ploidy Does Not Cause Genome-Wide Changes in DNA Methylation
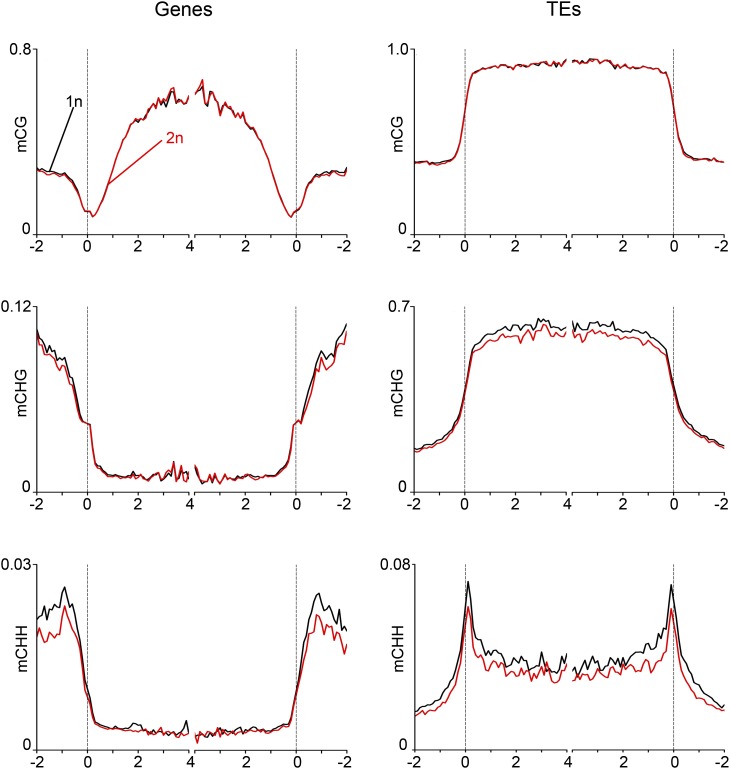
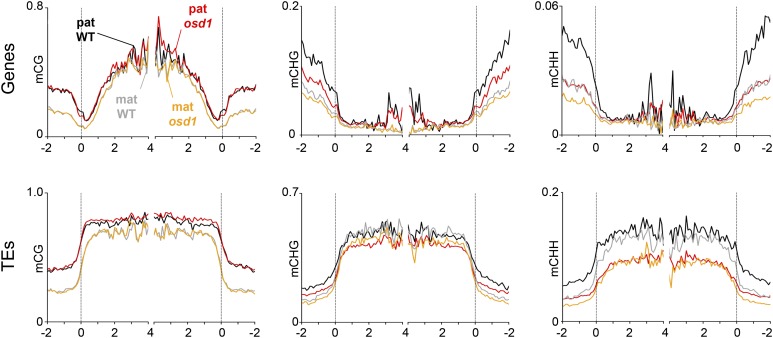
Polyploidization in Arabidopsis thaliana causes changes in DNA methylation (Mittelsten Scheid et al., 2003; Yu et al., 2010; Donoghue et al., 2014); however, when these changes are established remains to be shown. We addressed the question whether increased paternal genome dosage causes altered DNA methylation patterns in sperm that in turn cause global changes of gene expression after fertilization. The meiotic mutant jason (jas) forms diploid pollen containing 2n sperm cells at ∼60% frequency (Erilova et al., 2009; De Storme and Geelen, 2011), allowing the simultaneous isolation of 1n and 2n sperm cells. We isolated 1n and 2n sperm nuclei of jas-3 pollen by fluorescence-activated cell sorting and analyzed genome-wide DNA methylation profiles of 1n and 2n sperm genomes by shotgun bisulfite sequencing. There was no global change of CG methylation on genes and neither on TEs, while CHG and CHH methylation levels were slightly reduced in 2n compared with 1n sperm; however, those changes were not statistically significant (Figure 3). We thus conclude that increased paternal genome dosage does not cause global DNA methylation changes in sperm, suggesting that global deregulation of gene expression in triploid paternal excess seeds is not a consequence of DNA methylation changes in sperm.
Figure 3.
DNA Methylation Profile of 1n and 2n Sperm Nuclei.
Genes (left panels) and TEs (right panels) were aligned at the 5′ and 3′ ends (dashed lines), and average methylation levels in CG (top panels), CHG (middle panels), and CHH context (bottom panels) for each 100-bp interval were plotted.
Genome-Wide DNA Methylation Changes in Embryo and Endosperm of Triploid Paternal Excess Seeds
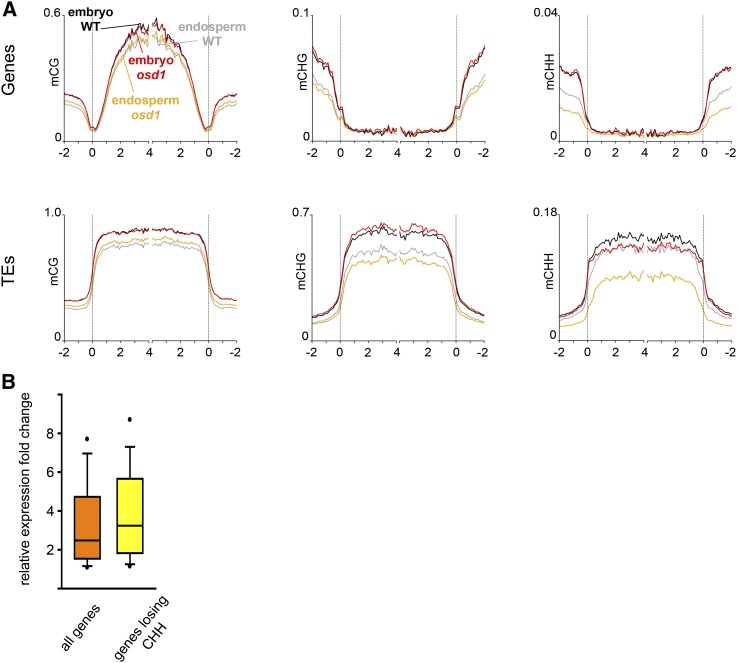
Decreased levels of 24-nucleotide small interfering RNAs were recently shown to correlate with deregulated gene expression in triploid paternal excess seeds (Lu et al., 2012). However, it has not been tested whether paternal excess indeed induces genome-wide de novo DNA methylation changes. We tested this hypothesis by generating allele-specific genome-wide DNA methylation profiles of embryo and endosperm in response to paternal excess interploidy hybridizations. We pollinated plants of the Ler accession with either wild-type (Col) or osd1-derived 2n pollen and dissected seeds at 7 DAP into embryo and endosperm fractions that were analyzed using shotgun bisulfite sequencing (Supplemental Table 1). Genome-wide DNA methylation profiles revealed that the endosperm of triploid seeds (4n endosperm) had strongly decreased CHH methylation levels at TEs (Kolmogorov-Smirnov test, P < 0.001) and flanking regions of genes (P value = 0.003) compared with wild-type 3n endosperm (Figure 4A). CHG methylation levels at TEs were also significantly reduced in the 4n endosperm (Kolmogorov-Smirnov test, P < 0.001), while CG methylation was slightly but significantly increased (Kolmogorov-Smirnov test, P < 0.001; Figure 4A). A minor reduction of CHH methylation was also observed within TEs of triploid embryos compared with 2n embryos (Figure 4A). We tested whether the loss of CHH methylation within the gene or gene flanking region (within 1 kb up- or downstream of ATG and stop codon, respectively; Supplemental Data Set 2) in the 4n endosperm would correlate with increased transcript levels. Indeed, genes with reduced CHH methylation levels were stronger upregulated in triploid seeds compared with genes that had no CHH methylation changes (Wilcoxon rank-sum test, P < 0.05; Figure 4B).
Figure 4.
DNA Methylation Changes in Embryo and Endosperm Derived from Interploidy Hybridizations and Effects on Transcript Levels.
(A) Genes (top panels) and TEs (lower panels) were aligned at the 5′ and 3′ ends (dashed lines), and average methylation levels in CG (left panels), CHG (middle panels), and CHH context (right panels) for each 100-bp interval were plotted.
(B) Box plots of expression changes in triploid seeds. The orange box shows expression changes of all genes with increased expression in triploid seeds, while the yellow box shows expression changes of those genes that have increased expression in triploid seeds but lose CHH methylation.
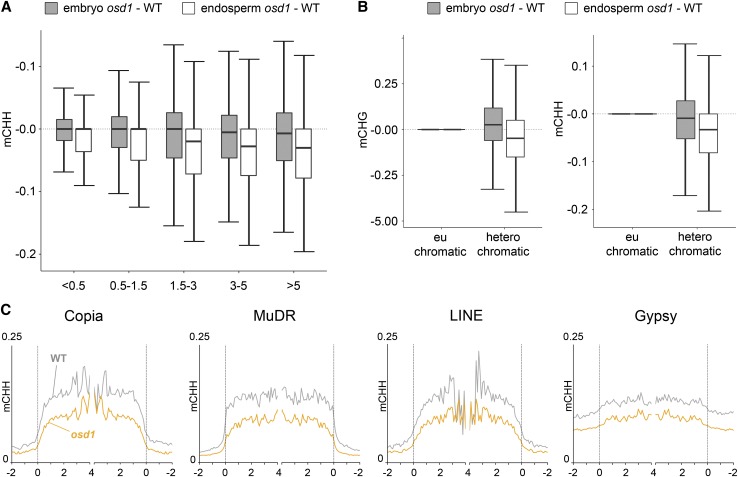
Making use of parental-specific single-nucleotide polymorphisms between Ler and Col accessions, we assessed which parental allele is affected by changes in DNA methylation in the endosperm. Reduced levels of CHH and CHG methylation at TEs in the 4n endosperm were detected on both maternal and paternal alleles (Kolmogorov-Smirnov test, P < 0.001for all tests; Figure 5). A particularly strong reduction of CHH methylation was detected at the paternal allele of gene-flanking regions in the 4n endosperm (Figure 5), revealing that maternal and paternal alleles can be differentially affected in the 4n endosperm. We tested whether loss of CHH methylation would correlate with TE length or TE location (euchromatic versus heterochromatic TEs). There was a clear trend of short euchromatic (those with low H3K9m2; Zemach et al., 2013) TEs being less susceptible to loss of CHH methylation in the endosperm, compared with H3K9m2-rich heterochromatic TEs (Figures 6A and 6B). We also tested whether different families of TEs would behave differently with respect to loss of CHH methylation; however, we did not detect a noticeable preference for a particular TE family (Figure 6C).
Figure 5.
Allele-Specific DNA Methylation Profiles of Endosperm Derived from Interploidy Hybridizations.
Genes (top panel) and TEs (lower panel) were aligned at the 5′ and 3′ ends (dashed lines), and average methylation levels in CG (left panels), CHG (middle panels), and CHH context (right panels) for maternal and paternal alleles for each 100-bp interval were plotted.
Figure 6.
DNA Methylation Differences Are Prominent on Long Heterochromatic TEs.
(A) Box plots showing absolute fractional CHH demethylation of 50-bp windows within transposons; numbers on x axis represent length of TEs in kilobases.
(B) Box plots showing absolute fractional CHG (left panel) and CHH (right panel) demethylation of 50-bp windows within euchromatic and heterochromatic TEs. TEs with H3K9m2 log2 scores lower than −1 and higher than 1 are considered euchromatic and heterochromatic, respectively.
(C) CHH methylation levels are plotted as in Figure 4 for TEs belonging to four distinct families.
Together, these data reveal that there is a postfertilization reduction of CHH methylation in the 4n endosperm that affects TEs and gene flanking regions and positively correlates with transcript levels.
Suppression of the Triploid Block by met1-Induced Paternal Genome Hypomethylation Does Not Require Maternal FIS-PRC2
Increased paternal genome dosage has been shown to cause preferential deregulation of target genes of FIS-PRC2 (Erilova et al., 2009; Tiwari et al., 2010). Consistent with these data, we found strong enrichment of H3K27m3 on genes showing increased expression levels in triploid seeds compared with diploid seeds (Figure 7A). Loss of FIS-PRC2 function in the endosperm causes strikingly similar changes of DNA methylation, as we report in this study: reduced levels of CHG and CHH methylation and increased levels of CG methylation (Ibarra et al., 2012). Previous studies revealed that hypomethylated pollen derived from a MET1 antisense plant can bypass maternal FIS-PRC2 requirement (Luo et al., 2000; Vinkenoog et al., 2000). Functional bypass of FIS-PRC2-specific subunits MEA and FIS2 occurs independently of a functional paternal MEA or FIS2 allele, revealing that reactivation of the paternal MEA and FIS2 alleles in met1 pollen cannot account for met1-mediated seed rescue (Luo et al., 2000). We tested whether hypomethylated pollen of a met1 mutant could bypass the maternal FIS-PRC2 requirement in 3n seeds by pollinating fis2 mutants with osd1 met1-derived 2n pollen. The majority of seeds derived from this cross were viable revealing that rescue of 3n seeds by hypomethylated pollen did not require a functional FIS-PRC2 (Figure 7B). Similar to the effect of met1 pollen on PEG expression in 3n seeds, met1 pollen could suppress deregulated expression of PEGs in 3n fis2 mutant seeds (Figure 7C). In agreement with a previous study (Vinkenoog et al., 2000), pollinations of fis2 with met1 pollen gave rise to two populations of seeds, one with decreased one with increased size, likely reflecting seeds carrying a wild-type and a mutant FIS2 allele, respectively (Supplemental Figure 1B). Pollinations of fis2 with osd1 met1 pollen gave rise to seeds that were substantially larger than seeds of fis2 × met1 pollinations and substantially larger than wild-type seeds (Supplemental Figure 1B), revealing the potential of epigenetic modifications for manipulating seed size.
Figure 7.
Rescue of 3n Seeds by Hypomethylated Pollen Does Not Require a Functional FIS-PRC2.
(A) H3K27m3 levels in the endosperm (data from Weinhofer et al., 2010) of genes upregulated (green line) and downregulated (blue line) in wild type × osd1 crosses compared with all genes (red line). Average methylation level is shown for 100-bp windows; numbers on x axis represent the length of genes in kilobases.
(B) Percentage of collapsed and partially collapsed seeds in interploidy hybridizations with lack of FIS-PRC2; numbers on top of bars represent total number of seeds per genotype.
(C) Expression analyses of PEGs in whole siliques 6 DAP derived from indicated crosses. MNE, mean normalized expression; ACTIN11 was used as reference. Error bars indicate se.
Suppression of the Triploid Block by met1 Pollen Is Likely Caused by Redistribution of DNA Methylation Marks in the Endosperm
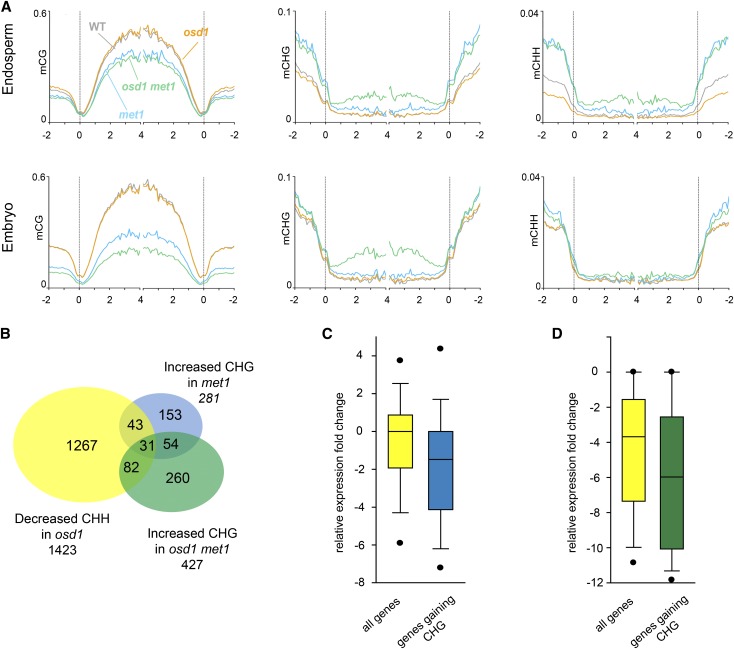
In the met1 mutant, CHG methylation becomes redistributed to euchromatic regions of chromosomes (Cokus et al., 2008; Lister et al., 2008; Stroud et al., 2013). We therefore reasoned that met1-induced suppression of triploid seed abortion might be a consequence of de novo CHG methylation of FIS-PRC2 target genes. To test this, we generated DNA methylation profiles of embryo and endosperm derived from 6 DAP seeds of the Ler accession pollinated with either met1 or osd1 met1-derived pollen. Methylation at CG residues in genes and TEs was strongly reduced in embryo and endosperm (Figure 8A; Supplemental Figure 4), revealing that paternal genome hypomethylation is not instantly restored after fertilization. Supporting the idea that hypomethylation causes redistribution of CHG methylation to genic regions, CHG methylation levels were strongly increased in genic regions in the endosperm derived from osd1 met1 pollinations (Kolmogorov-Smirnov test, P < 0.001) and moderately increased in the endosperm derived from met1 pollinations (Kolmogorov-Smirnov test, P < 0.001; Figure 8A). Genes with increased CHG methylation levels upon met1 and osd1 met1 pollinations significantly overlapped with genes losing CHH in triploid seeds (Hypergeometric test, P = 4.62E-25 and P = 4.62E-57, respectively; Figure 8B). Importantly, gain of CHG methylation upon pollination with met1 and osd1 met1 pollen correlated with decreased gene expression (Wilcoxon rank-sum test, P < 0.05; Figures 8C and 8D; Supplemental Data Set 2). Furthermore, many PEGs that had increased expression levels in osd1 derived seeds and were transcriptionally suppressed by osd1 met1 pollinations gained CHG methylation within the gene body or flanking regions after osd1 met1 pollinations compared with osd1 pollinations (Supplemental Figure 5). In the endosperm derived from osd1 met1 pollinations, there was also a marked increase of CHH methylation in genic regions and TEs compared with endosperm from osd1 pollinations (Kolmogorov-Smirnov test, P < 0.001 for both tests), restoring wild-type CHH methylation levels at TEs (Figure 8A; Supplemental Figure 4). Together, these data reveal that met1-induced paternal hypomethylation causes redistribution of CHG and CHH methylation marks to genic regions in the 4n endosperm, which negatively impacts on gene expression.
Figure 8.
Loss of CG Methylation Causes Redistribution of CHG and CHH Methylation.
(A) DNA methylation profiles of genes in the endosperm (top panels) and embryo (lower panels) derived from seeds of the following crosses: wild type × wild type, wild type × osd1, wild type × osd1 met1, and wild type × met1. Genes were aligned at the 5′ and 3′ends (dashed lines), and average methylation levels in CG (left panels), CHG (middle panels), and CHH context (right panels) for maternal and paternal alleles for each 100-bp interval were plotted.
(B) Venn diagram showing overlap of genes with decreased CHH methylation in wild type × osd1 crosses compared with the wild type with genes that have increased CHG methylation levels in wild type × met1 and wild type × osd1 met1 compared with wild type and wild type × osd1, respectively.
(C) Box plots of expression changes in diploid seeds upon pollination with met1 pollen compared with wild-type pollinations. The yellow box shows expression changes of all genes with increased expression in triploid seeds, while the blue box shows expression changes of genes that gain CHG methylation upon met1 pollination.
(D) Box plots of expression changes in triploid seeds upon pollination with osd1 met1 pollen compared with osd1 pollinations. The yellow box shows expression changes of all genes with increased expression in triploid seeds, while the green box shows expression changes of genes that gain CHG methylation upon osd1 met1 pollination.
DISCUSSION
In this study, we uncover an epigenetic mechanism that allows bypassing the postzygotic interploidy hybridization barrier in Arabidopsis, leading to the formation of viable triploid seeds. Our study reports four major findings: (1) met1-induced hypomethylation in pollen can suppress triploid seed abortion; (2) meiotic defects leading to the formation of unreduced pollen do not cause major DNA methylation changes in sperm cells; (3) the tetraploid endosperm of paternal excess seeds has strongly reduced levels of CHG and CHH methylation; and (4) bypass of triploid seed abortion by met1 pollen is likely a consequence of de novo CHG methylation targeted to FIS-PRC2 target genes.
Triploid paternal excess seeds have phenotypic defects and gene expression profiles similar to seeds lacking FIS-PRC2 function (Erilova et al., 2009; Tiwari et al., 2010), suggesting that FIS-PRC2 function is affected in triploid seeds. However, our data reveal that met1-mediated triploid seed rescue is not caused by restoring FIS-PRC2 function, but acts via a different mechanism. Similarly, hypomethylated pollen can rescue mutants impaired in FIS-PRC2 function (Vielle-Calzada et al., 1999; Luo et al., 2000; Vinkenoog et al., 2000), strongly suggesting that triploid seed rescue and fis mutant rescue by hypomethylated met1 pollen have a similar mechanistic basis. Rescue of fis2 and mea mutants by hypomethylated pollen does not require functional paternal FIS2 or MEA alleles (Luo et al., 2000), suggesting that paternal CG hypomethylation activates a repressive pathway allowing bypass of FIS-PRC2 function. We show that in the CG hypomethylated tetraploid endosperm, CHG methylation was redistributed to genic regions, which likely accounts for gene repression in the absence of FIS-PRC2 function. Supporting this proposition, redistribution of CHG methylation to euchromatic regions in met1 mutant plants was previously reported and correlated with gene repression (Lister et al., 2008). Similarly, we observed a negative correlation of de novo recruited CHG methylation and gene expression. Mutations in the JmjC H3K9m2 demethylase INCREASE IN BONSAI METHYLATION1 (IBM1) gene cause increased CHG methylation within gene bodies, which is associated with severe developmental abnormalities (Miura et al., 2009). Expression of IBM1 was not significantly changed in hypomethylated diploid and triploid seeds (Supplemental Data Set 1), making it unlikely that reduced IBM1 function is causally responsible for increased CHG methylation in hypomethylated seeds. Supporting this notion, increased H3K9m2 levels in the met1 mutant are not dependent on ibm1 (Deleris et al., 2012), revealing an as yet unknown targeting signal responsible for de novo recruitment of CHG methylation in the met1 mutant. Importantly, H3K27m3 and H3K9m2/ CHG DNA methylation were suggested to replace one another in a locus-specific manner (Deleris et al., 2012). Thus, there is precedence for our proposition that gain of CHG methylation upon met1 pollination can suppress genes that become upregulated in triploid seeds. Those deregulated genes are preferentially marked by H3K27m3 in wild-type seeds, suggesting that in the endosperm, like in vegetative tissues, CHG methylation can suppress PRC2 target genes.
While previous research revealed that polyploidization causes changes in DNA methylation (Mittelsten Scheid et al., 2003; Yu et al., 2010; Donoghue et al., 2014), the time point when these changes are established is unclear. Our data demonstrate that meiotic defects leading to nuclear restitution and unreduced pollen formation do not induce detectable changes in DNA methylation, revealing that the triploid response is not a consequence of genome-wide changes of DNA methylation in unreduced pollen. Therefore, DNA methylation changes in triploid seeds are established after fertilization. Accordingly, genome-wide reduction of CHG and CHH methylation levels in the tetraploid endosperm occurred at TEs of both parental genomes. We found that the most preferred targets for the loss of CHH methylation were long heterochromatic TEs. Pericentromeric TEs were shown to become strongly depleted of CHH methylation in sperm (Calarco et al., 2012), suggesting that the loss of CHH methylation in triploid seeds is a consequence of twice as many CHH depleted paternal genomes. It has been proposed that after fertilization, CHH methylation in the endosperm is guided by maternally produced small 24-nucleotide RNAs (Mosher et al., 2009; Calarco et al., 2012; Lu et al., 2012). Therefore, remethylation of two paternal genomes could be limited by insufficient numbers of maternal 24-nucleotide RNAs. This scenario is in agreement with the finding that 24-nucleotide small RNAs depend on maternal genome dosage, causing reduced levels of 24-nucleotide RNAs in triploid paternal excess seeds (Lu et al., 2012).
The paternally expressed imprinted gene ADM has been shown to build the interploidy hybridization barrier in Arabidopsis (Kradolfer et al., 2013b). ADM expression was suppressed upon met1 and osd1 met1 pollinations, suggesting that ADM could be one of the causal genes that restore viable triploid seed formation upon met1-mediated suppression. ADM is conserved in the closely related genera Arabidopsis and Capsella, but there are no ADM orthologs in more distantly related genera or outside the Brassicaceae. However, both within and outside the Brassicaceae, there are many species that are sensitive to interploidy hybridizations (Ramsey and Schemske, 1998), indicating that there are other causal targets in addition to ADM. Importantly, met1 pollen caused genome-wide normalization of gene expression in triploid seeds by reconfiguring DNA methylation marks that are likely to occur independently of ADM. This strongly suggests that triploid seed rescue by met1 is widely applicable to other species that are sensitive to interploidy hybridizations. Therefore, our findings are of major importance, as they may represent a generally applicable way to overcome interploidy hybridization barriers, opening new avenues to plant breeding.
METHODS
Plant Material and Growth Conditions
All seed material was surface sterilized (5% sodium hypochlorite and 0.01% Triton X-100), stratified for 2 d at 4°C and germinated on half-strength Murashige and Skoog medium (1% sucrose) in long-day conditions (16 h light/8 h darkness; 21°C). After 10 d in light conditions, germination ratios were determined and/or plants were transferred to soil and grown under long day conditions.
Arabidopsis thaliana mutants jas-3 (Erilova et al., 2009), fis2-5 (Weinhofer et al., 2010), and met1-3 (Saze et al., 2003) are in the Col-0 accession. The osd1-1 mutant (d’Erfurth et al., 2009) was kindly provided by Raphael Mercier. The mutant was originally identified in the Nossen background and subsequently introgressed into Col by repeated backcrossing over five generations. The osd1-1 met1-3 double mutant was generated by crossing heterozygous individuals of both single mutants. Seed stocks of Col-0 2x (N1093) and its colchicine-derived autotetraploid (Col 4X) were kindly donated by L. Comai (University of California, Davis). met1-9 (Tiwari et al., 2010) was treated with colchicine to obtain 4x met1-9 (Supplemental Methods).
Generation of Endosperm and Embryo Tissue Material
Ler plants were emasculated and after 2 d of pollination with Col, osd1-1, osd1-1 met1-3, or met1-3 pollen. The 6- to 7-DAP seeds were dissected on a glass slide into endosperm and embryo samples in 0.3 M sorbitol/5 mM MES solution under a stereoscope using fine needles. Seed coat tissue was discarded and embryo/endosperm material was washed in sorbitol solution. Supernatant was removed and samples were frozen in liquid nitrogen and stored until all material was collected. Quality of isolated embryo and endosperm fractions was tested by calculating the maternal to paternal genome ratio after bisulfite sequencing (Supplemental Table 1).
RNA Sequencing and Expression Analysis
To obtain material for RNA sequencing seeds from 20 siliques of Ler × Col, Ler × osd1-1, Ler × osd1-1 met1-3, and Ler × met1-3 crosses were harvested into RNA later (Sigma-Aldrich) in triplicate and homogenized (Silamat S5; IvoclarVivadent) using glass beads. RNA was extracted following a modified protocol for the RNAqueous kit (Ambion, Life Technologies), and residual DNA was removed using 5 μL DNaseI (Thermo-Scientific), followed by phenol-chloroform extraction and ethanol precipitation. RNA was sequenced at the SciLife Laboratory (Uppsala, Sweden) on an Illumina HiSeq2000 on two lanes in 100-bp paired-end fashion. Sequencing reads are deposited as fastq files in the Gene Expression Omnibus (GSE53642).
Seeds used for expression analysis were collected from 20 siliques at 4 and 6 DAP. RNA from whole seeds was obtained as described above. RNA from endosperm and embryo material for expression analysis was extracted from seeds of 30 siliques using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions, and residual DNA was removed using the Qiagen RNase-free DNase set. RNA of siliques was extracted using the RNeasy Plant Mini Kit according to the manufacturer’s instructions, and residual DNA was removed using the Qiagen RNase-free DNase set. cDNA for all RNA samples was synthesized using the first-strand cDNA synthesis kit (Fermentas) according to the manufacturer’s instructions.
Quantitative real-time PCR was performed using an iQ5 Real-Time PCR detection system (Bio-Rad) in triplicates using Maxima SYBR green master mix (Fermentas). Results were analyzed as described by Simon (2003) using ACTIN11 as a reference gene (for primer sequences, see Supplemental Table 2).
DNA Extraction and Bisulfite Conversion
To obtain material for bisulfite sequencing endosperm and embryo material from ∼60 siliques of Ler × Col, Ler × osd1-1, Ler × osd1-1 met1-3, and Ler × met1-3 crosses was collected over several days and stored at −70°C before it was finally combined for DNA extraction. DNA was extracted using 2xCTAB (2% [w/v] CTAB, 1.4 M NaCl, 20 mM EDTA, and 100 mM Tris-HCl, pH 8), followed by phenol/chloroform extraction and subsequent ethanol precipitation.
Bisulfite Library Preparation
After DNA extraction using 2xCTAB, concentrations were measured using Qubit (Life Technologies), and all material (50 to 200 ng DNA/sample, depending on the tissue) was subjected to bisulfite conversion and library preparation. DNA shearing was performed in 80 μL for 30 cycles (30 s on; 30 s off) using Bioruptor NextGen (Diagenode). Bisulfite conversion was performed using the Epitect kit (Qiagen) following the manufacturer’s instructions. The library was prepared using the Illumina TrueSeq DNA LT sample preparation kit.
High-Throughput RNA Sequence Analysis
Sequencing reads were aligned to the TAIR 10.0 version of the Arabidopsis reference genome (Col-0) using TopHat (Trapnell et al., 2009). Only uniquely mapping reads were used for further analysis. Differentially expressed genes were identified using the DESeq package (Anders and Huber, 2010). For assigning reads as Ler or Col, we used the publically available Samtools package (http://sourceforge.net/projects/samtools/files/samtools/0.1.18/samtools-0.1.18.tar.bz2/download). Ler single-nucleotide polymorphisms were downloaded from the 1001 Genomes Data Center (http://1001genomes.org/data/MPI/MPISchneeberger2011/releases/current/Ler-1/Marker/Ler-1.SNPs.TAIR9.txt). The allele counts per gene were summed for the triplicates. We analyzed MEGs and PEGs commonly identified in Col and Ler accessions (Gehring et al., 2011; Hsieh et al., 2011) and uniquely identified in Col and Bur accessions (Wolff et al., 2011) (Supplemental Data Set 1). Sequencing data are deposited in the Gene Expression Omnibus (GSE53642).
Bisulfite Sequence Analysis
Reads from the Illumina BS sequence libraries were aligned to the TAIR10 Arabidopsis genome using the Bismark read mapper (Krueger and Andrews, 2011) allowing up to two mismatches per read. Duplicated read pairs (aligning to the same genomic position) were eliminated before performing methylation analyses.
Methylation Analysis
TAIR10 annotated genes and transposons were aligned at the 5′ and 3′ end (dashed lines), and average methylation levels for each 100-bp interval were plotted from 2 kb away from the genetic feature (negative numbers) to 4 kb into the genetic feature (positive numbers). Only positions with at least five informative cytosines were considered. We discarded from the analysis half of the length of the genetic feature (from the end opposite to the one used in the alignment) in order to avoid averaging terminal and central regions in genes and transposons having different length. Parental-of-origin (allele specific) methylation plots were constructed using reads that could be unequivocally assigned to one of the parents (reads aligning without mismatches and to either the Col or the Ler genomes). Differentially methylated regions were identified as previously described (Ibarra et al., 2012), and genes containing differentially methylated regions are indicated in Supplemental Data Set 2. A two-sample Kolmogorov-Smirnov test was applied to test for significant methylation differences in 100-bp windows across either flanking regions or bodies of genes and transposable elements.
Box Plots
Box plots show absolute fractional demethylation of 50-bp windows within transposons. The height of the box encloses the middle 50% of the distribution and the horizontal line marks the median. Vertical lines delimitate the minimum and maximum values that fall within 1.5 times the height of the box. Only positions with at least five informative cytosines were considered.
Isolation of Sperm Nuclei and DNA Extraction
Isolation of pollen nuclei was described previously (Schoft et al., 2011). Briefly, pollen was obtained by repeated washes of collected inflorescences with 9% sucrose. Pollen grains were transferred to buffer A (1 M sorbitol, 7% Ficol PM 400, 5 mM MgAc, 3 mM CaCl2, 5 mM EGTA, 50 mM Tris-HCl, pH 7.5, 20% glycerol, 2% Triton X-100, protease inhibitor cocktail, and 1 mM final concentration phenylmethylsulfonyl fluoride) and disrupted by vortexing with glass beads. Following dilution with buffer B (15 mM Tris-HCl, pH 7.5, 2 mM Na2-EDTA, 0.5 mM spermine-4 HCl, 80 mM KCl, 20 mM NaCl, 2% Triton X-100, protease inhibitor cocktail, and phenylmethylsulfonyl fluoride), free pollen nuclei were sorted by fluorescence-activated cell sorting based on differences in SYBR green I staining using a BD FACSAria I device (Becton Dickinson). Four populations of jas-3 pollen nuclei can be detected by SYBR green I, and 1n and 2n sperm nuclei were selected for sorting. Purity of the sorted nuclei populations was confirmed by 4′,6-diamidino-2-phenylindole staining and scored by fluorescence microscopy.
DNA was isolated from the sorted 1n and 2n sperm cell nuclei by proteinase K digestion and subsequent phenol extraction. DNA samples from several sorts were pooled, washed with distilled water, and concentrated with Amicon Ultra-4 (Ultracel-50k) centrifugal filter devices (Millipore).
Accession Numbers
The accession numbers for the genes discussed in this article are as follows: ADM (AT4G11940), AGL28 (AT1G01530), AGL36 (AT5G26650), AGL40 (AT4G36590), AGL42 (AT5G62165), AGL48 (AT2G40210), AGL62 (AT5G60440), AGL90 (AT5G27960), AGL96 (AT5G06500), FIS2 (AT2G35670), IKU1 (AT2G35230), IKU2 (AT3G19700), MEA (AT1G02580), NPR1(AT1G64280), PHE1 (AT1G65330), PKR2 (AT4G31900), SID2 (AT1G74710), SUVH7 (AT1G17770), TT2 (AT5G35550), and YUC10 (AT1G48910). The transcriptome sequencing data and bisulfite sequencing data have been submitted to the National Center for Biotechnology Information’s Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/) and are available under accession number GSE53642.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of Seed Size from Indicated Crosses.
Supplemental Figure 2. Tetraploid met1 Can Bypass the Triploid Block.
Supplemental Figure 3. Fold Change Expression of Selected Genes in Triploid Seeds and Hypomethylated Diploid and Triploid Seeds.
Supplemental Figure 4. Methylation Profiles of Transposable Elements (TEs) in the Embryo and Endosperm after Pollination with Hypomethylated Pollen.
Supplemental Figure 5. CHG Methylation Profiles of PEGs in the Endosperm.
Supplemental Table 1. Median Coverage per Cytosine and Mean DNA Methylation for Samples Analyzed in This Study.
Supplemental Table 2. List of Primers Used in Quantitative RT-PCR.
Supplemental Methods and References.
Supplemental Data Set 1. mRNA levels, Expression Fold Changes, P Values, and Allele-Specific Read Counts for Samples Ler × Col, Ler × osd1, Ler × osd1 met1, and Ler × met1.
Supplemental Data Set 2. Genes Containing Differentially Methylated Regions within the Gene or in Gene-Flanking Regions (1 kb Up- or Downstream or ATG or Stop Codon, Respectively).
Supplementary Material
Acknowledgments
We thank Cecilia Wärdig for excellent technical support. Sequencing was performed by the SNP&SEQ Technology Platform, Science for Life Laboratory at Uppsala University, a national infrastructure supported by the Swedish Research Council (VRRFI) and the Knut and Alice Wallenberg Foundation. This research was supported by a European Research Council Starting Independent Researcher grant (to C.K.), a grant from the Swedish Science Foundation (to C.K.), and a grant from the Knut and Alice Wallenberg Foundation (to C.K.).
AUTHOR CONTRIBUTIONS
C.K., N.S., P.W., and R.S. designed the research. N.S., P.W., V.S., and R.S. performed research. C.K., N.S., P.W., H.T., R.S., J.S.-G., and A.S. analyzed data. C.K. and N.S. wrote the article and all authors contributed to the editing.
Glossary
- TE
transposable element
- Col
Columbia
- DAP
days after pollination
- Ler
Landsberg erecta
- MEG
maternally expressed gene
- PEG
paternally expressed gene
- SLR
signal log ratio
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Adams, S., Vinkenoog, R., Spielman, M., Dickinson, H.G., Scott, R.J. (2000). Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127: 2493–2502 [DOI] [PubMed] [Google Scholar]
- Anders, S., Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J.A. (1993). Dosage analysis of maize endosperm development. Annu. Rev. Genet. 27: 181–204 [DOI] [PubMed] [Google Scholar]
- Brink, R., Cooper, D. (1947). The endosperm in seed development. Bot. Rev. 132: 423–541 [Google Scholar]
- Burkart-Waco, D., Ngo, K., Dilkes, B., Josefsson, C., Comai, L. (2013). Early disruption of maternal-zygotic interaction and activation of defense-like responses in Arabidopsis interspecific crosses. Plant Cell 25: 2037–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco, J.P., Borges, F., Donoghue, M.T., Van Ex, F., Jullien, P.E., Lopes, T., Gardner, R., Berger, F., Feijó, J.A., Becker, J.D., Martienssen, R.A. (2012). Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus, S.J., Feng, S., Zhang, X., Chen, Z., Merriman, B., Haudenschild, C.D., Pradhan, S., Nelson, S.F., Pellegrini, M., Jacobsen, S.E. (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris, A., Stroud, H., Bernatavichute, Y., Johnson, E., Klein, G., Schubert, D., Jacobsen, S.E. (2012). Loss of the DNA methyltransferase MET1 Induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet. 8: e1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Erfurth, I., Jolivet, S., Froger, N., Catrice, O., Novatchkova, M., Mercier, R. (2009). Turning meiosis into mitosis. PLoS Biol. 7: e1000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme, N., Geelen, D. (2011). The Arabidopsis mutant jason produces unreduced first division restitution male gametes through a parallel/fused spindle mechanism in meiosis II. Plant Physiol. 155: 1403–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue, M.T., Fort, A., Clifton, R., Zhang, X., McKeown, P.C., Voigt-Zielinksi, M.L., Borevitz, J.O., Spillane, C. (2014). C(m)CGG methylation-independent parent-of-origin effects on genome-wide transcript levels in isogenic reciprocal F1 triploid plants. DNA Res. 21: 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G.N., Yadegari, R. (2002). Development and function of the angiosperm female gametophyte. Annu. Rev. Genet. 36: 99–124 [DOI] [PubMed] [Google Scholar]
- Erilova, A., Brownfield, L., Exner, V., Rosa, M., Twell, D., Mittelsten Scheid, O., Hennig, L., Köhler, C. (2009). Imprinting of the polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLoS Genet. 5: e1000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, M., Bubb, K.L., Henikoff, S. (2009). Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324: 1447–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, M., Missirian, V., Henikoff, S. (2011). Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS ONE 6: e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Marcos, J.F., Pennington, P.D., Costa, L.M., Dickinson, H.G. (2003). Imprinting in the endosperm: a possible role in preventing wide hybridization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig, D., Westoby, M. (1989). Parent specific gene expression and the triploid endosperm. Am. Nat. 134: 147–155 [Google Scholar]
- Hehenberger, E., Kradolfer, D., Köhler, C. (2012). Endosperm cellularization defines an important developmental transition for embryo development. Development 139: 2031–2039 [DOI] [PubMed] [Google Scholar]
- Hsieh, T.F., Ibarra, C.A., Silva, P., Zemach, A., Eshed-Williams, L., Fischer, R.L., Zilberman, D. (2009). Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, T.F., Shin, J., Uzawa, R., Silva, P., Cohen, S., Bauer, M.J., Hashimoto, M., Kirkbride, R.C., Harada, J.J., Zilberman, D., Fischer, R.L. (2011). Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. USA 108: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra, C.A., et al. (2012). Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, J.P., Lindroth, A.M., Cao, X., Jacobsen, S.E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Kinoshita, T. (2007). Reproductive barrier and genomic imprinting in the endosperm of flowering plants. Genes Genet. Syst. 82: 177–186 [DOI] [PubMed] [Google Scholar]
- Kradolfer, D., Hennig, L., Köhler, C. (2013a). Increased maternal genome dosage bypasses the requirement of the FIS polycomb repressive complex 2 in Arabidopsis seed development. PLoS Genet. 9: e1003163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kradolfer, D., Wolff, P., Jiang, H., Siretskiy, A., Köhler, C. (2013b). An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev. Cell 26: 525–535 [DOI] [PubMed] [Google Scholar]
- Krueger, F., Andrews, S.R. (2011). Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, J.A., Jacobsen, S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Berger, F. (2012). Endosperm: food for humankind and fodder for scientific discoveries. New Phytol. 195: 290–305 [DOI] [PubMed] [Google Scholar]
- Lindroth, A.M., Cao, X., Jackson, J.P., Zilberman, D., McCallum, C.M., Henikoff, S., Jacobsen, S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Lister, R., O’Malley, R.C., Tonti-Filippini, J., Gregory, B.D., Berry, C.C., Millar, A.H., Ecker, J.R. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J., Zhang, C., Baulcombe, D.C., Chen, Z.J. (2012). Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 109: 5529–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M., Bilodeau, P., Dennis, E.S., Peacock, W.J., Chaudhury, A. (2000). Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, G. (1966). The origin and significance of intraspecific polyploidy: experimental evidence from Solanum chacoense. Evolution 20: 552–557 [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid, O., Afsar, K., Paszkowski, J. (2003). Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat. Genet. 34: 450–454 [DOI] [PubMed] [Google Scholar]
- Miura, A., Nakamura, M., Inagaki, S., Kobayashi, A., Saze, H., Kakutani, T. (2009). An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J. 28: 1078–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher, R.A., Melnyk, C.W., Kelly, K.A., Dunn, R.M., Studholme, D.J., Baulcombe, D.C. (2009). Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286 [DOI] [PubMed] [Google Scholar]
- Otto, S.P., Whitton, J. (2000). Polyploid incidence and evolution. Annu. Rev. Genet. 34: 401–437 [DOI] [PubMed] [Google Scholar]
- Pignatta, D., Gehring, M. (2012). Imprinting meets genomics: new insights and new challenges. Curr. Opin. Plant Biol. 15: 530–535 [DOI] [PubMed] [Google Scholar]
- Ramsey, J., Schemske, D. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29: 467–501 [Google Scholar]
- Saze, H., Mittelsten Scheid, O., Paszkowski, J. (2003). Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34: 65–69 [DOI] [PubMed] [Google Scholar]
- Schoft, V.K., Chumak, N., Choi, Y., Hannon, M., Garcia-Aguilar, M., Machlicova, A., Slusarz, L., Mosiolek, M., Park, J.S., Park, G.T., Fischer, R.L., Tamaru, H. (2011). Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl. Acad. Sci. USA 108: 8042–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, R.J., Spielman, M., Bailey, J., and Dickinson, H.G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329–3341 [DOI] [PubMed] [Google Scholar]
- Simon, J.A., Kingston, R.E. (2013). Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49: 808–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, P. (2003). Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19: 1439–1440 [DOI] [PubMed] [Google Scholar]
- Stroud, H., Do, T., Du, J., Zhong, X., Feng, S., Johnson, L., Patel, D.J., Jacobsen, S.E. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud, H., Greenberg, M.V., Feng, S., Bernatavichute, Y.V., Jacobsen, S.E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, S., Spielman, M., Schulz, R., Oakey, R.J., Kelsey, G., Salazar, A., Zhang, K., Pennell, R., Scott, R.J. (2010). Transcriptional profiles underlying parent-of-origin effects in seeds of Arabidopsis thaliana. BMC Plant Biol. 10: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C., Pachter, L., Salzberg, S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada, J.P., Thomas, J., Spillane, C., Coluccio, A., Hoeppner, M.A., Grossniklaus, U. (1999). Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev. 13: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinkenoog, R., Spielman, M., Adams, S., Fischer, R.L., Dickinson, H.G., Scott, R.J. (2000). Hypomethylation promotes autonomous endosperm development and rescues postfertilization lethality in fie mutants. Plant Cell 12: 2271–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhofer, I., Hehenberger, E., Roszak, P., Hennig, L., Köhler, C. (2010). H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 6: e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, P., Weinhofer, I., Seguin, J., Roszak, P., Beisel, C., Donoghue, M.T., Spillane, C., Nordborg, M., Rehmsmeier, M., Köhler, C. (2011). High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 7: e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, W., Brown, R.C., Lemmon, B.E., Harada, J.J., Goldberg, R.B., Fischer, R.L. (2006). Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol. 142: 1160–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z., Haberer, G., Matthes, M., Rattei, T., Mayer, K.F., Gierl, A., Torres-Ruiz, R.A. (2010). Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 17809–17814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach, A., Kim, M.Y., Hsieh, P.H., Coleman-Derr, D., Eshed-Williams, L., Thao, K., Harmer, S.L., Zilberman, D. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.