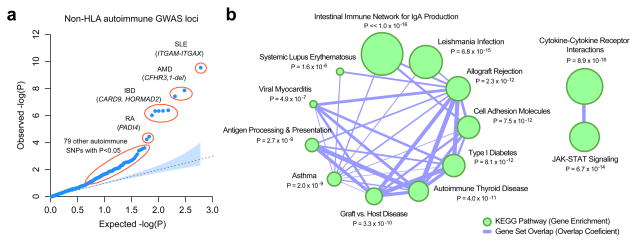
Figure 3. Autoimmunity/inflammatory loci and risk of IgAN.
(a) A quantile-quantile plot of IgAN associations for 582 unique non-HLA SNPs previously associated with autoimmune or immune-mediated diseases at p < 5 × 10−8 in the NHGRI GWAS catalogue. When tested for association with IgAN, an unexpectedly large number of SNPs deviate from the null expectation (empiric p < 1 × 10−4, Supplementary Figure 9). (b) The KEGG enrichment map for the genes residing within autoimmunity loci associated with IgAN at p < 0.05 (q < 0.25). The size of nodes reflects −log10-transformed P-values of the adjusted hypergeometric enrichment test in GSEA. The edges represent pathway similarity as defined by an overlap coefficient. The top overrepresented KEGG pathway is the “Intestinal Immune Network for IgA Production” (gene set overlap coefficient = 25%, enrichment p < 1.0 × 10−16). Individual genes intersecting top-ranked KEGG pathways are provided in Supplementary Figure 10c.