Introductory Paragraph
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoma subtype and is clinically aggressive. To identify genetic susceptibility loci for DLBCL, we conducted a meta-analysis of three new genome-wide association studies (GWAS) and one prior scan, totaling 3,857 cases and 7,666 controls of European ancestry, with additional genotyping of nine promising SNPs in 1,359 cases and 4,557 controls. In our multi-stage analysis, five independent SNPs in four loci achieved genome-wide significance marked by rs116446171 at 6p25.3 (EXOC2; P=2.33×10−21), rs2523607 at 6p21.33 (HLA-B; 2.40×10−10), rs79480871 at 2p23.3 (NCOA1; P=4.23×10−8), and two independent SNPs, rs13255292 and rs4733601, at 8q24.21 (PVT1; P=9.98×10−13 and P=3.63×10−11, respectively). These data provide substantial new evidence for genetic susceptibility to this B-cell malignancy, and point towards pathways involved in immune recognition and immune function in the pathogenesis of DLBCL.
Diffuse large B-cell lymphoma (DLBCL), the most common subtype of non-Hodgkin lymphoma (NHL)1, has an aggressive clinical course2. The risk of DLBCL is increased in individuals with a family history of NHL (odds ratio (OR)=1.4; 95%CI 1.1–2.0)3, supporting a genetic contribution. Also, relatives of DLBCL patients are at elevated risk for both DLBCL (RR=9.8, 95%CI 3.1–31) and Hodgkin lymphoma (HL, RR=2.0, 95%CI 1.05–4.0), but not indolent lymphomas4. Among candidate gene studies investigating susceptibility to DLBCL, only one locus, the LTA252G/TNF-308A haplotype on chromosome 6p21, reached genome-wide significance (P=2.9×10−8)5. In small GWAS of all NHL subtypes combined, no conclusive loci for NHL or DLBCL were identified in individuals of European background6–9, whereas a recent study conducted in East Asia identified a locus at 3q2710.
To discover new DLBCL susceptibility loci, in stage 1, we genotyped 2,878 DLBCL cases and 2,854 controls of European ancestry from 22 studies using the Illumina OmniExpress Beadchip (Online Methods; Supplementary Table 1; Supplementary Figure 1). A total of 5,346 (93.3%) samples and 611,844 SNPs successfully passed rigorous quality control criteria (Online Methods; Supplementary Table 2). To augment the number of controls, genotype data from 3,536 cancer-free controls previously analyzed with the Omni2.5 SNP microarray were folded into the analytical build11, resulting in a total of 2,661 cases and 6,221 controls for the stage 1 GWAS analysis (Supplementary Table 2).
In stage 1, with adjustment for gender, age and four eigenvectors (Online Methods), we observed an enrichment of SNPs with smaller P-values compared to the null distribution in the Q-Q plot with a lambda of 1.016 (Supplementary Figure 2). Two SNPs exceeded the threshold for genome-wide significance (P<5×10−8) whereas 20 SNPs showed highly suggestive associations (P<5×10−7) (Supplementary Figure 3). All but one SNP mapped to the HLA region of chromosome 6 (29.5Mb to 33.2Mb on Human Genome version 19 coordinates).
In stage 2, we included data from two unpublished and previously genotyped GWAS (GELA/EPIC and Mayo) plus one published GWAS (UCSF7), totaling 1,196 DLCBL cases and 1,445 controls (Online Methods; Supplementary Tables 1, 3). Because different genotyping platforms were used, we imputed common SNPs for each study based on the 1000 Genomes Project release version 312 and IMPUTE213 (Supplementary Table 4). In meta-analysis of all genotyped and high-quality imputed SNPs from stages 1 and 2 (N=8,363,971), we identified 19 SNPs at genome-wide significance (P<5×10−8) (Supplementary Table 5) and 134 SNPs at a suggestive level of significance (P<5×10−7) (Supplementary Table 6); 123 of the 153 total SNPs mapped to the HLA region on chromosome 6. Based on these results, we selected and successfully designed TaqMan primers for eight promising SNPs (P<5×10−6) outside the HLA region and one SNP from the HLA region for stage 3 de novo genotyping in an additional 1,359 DLBCL cases and 4,557 controls (Online Methods; Supplementary Tables 1, 3).
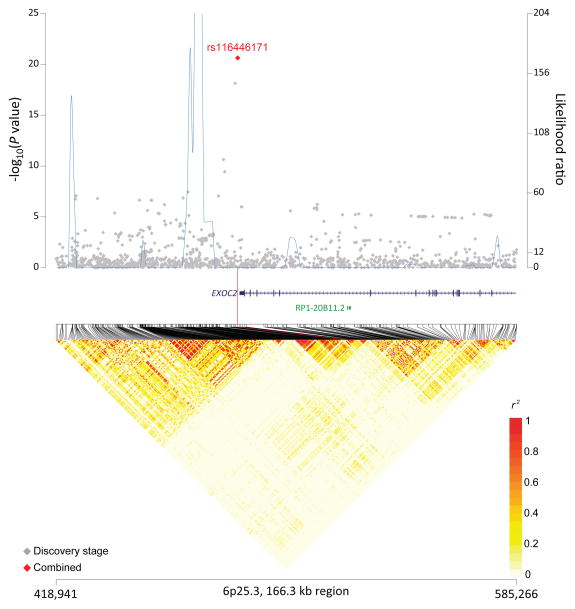
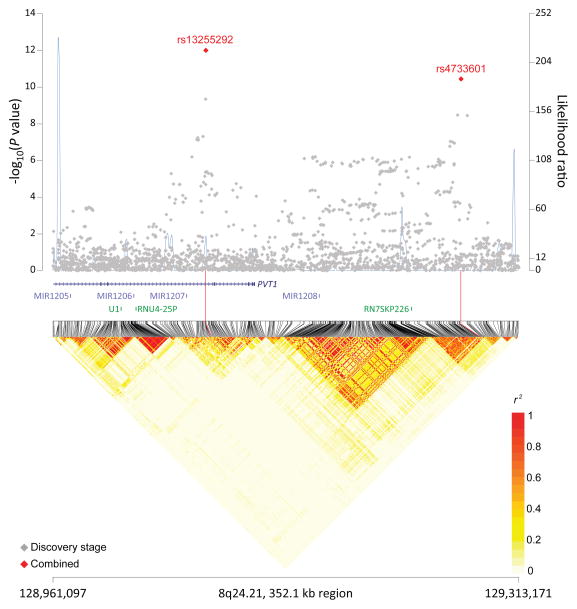
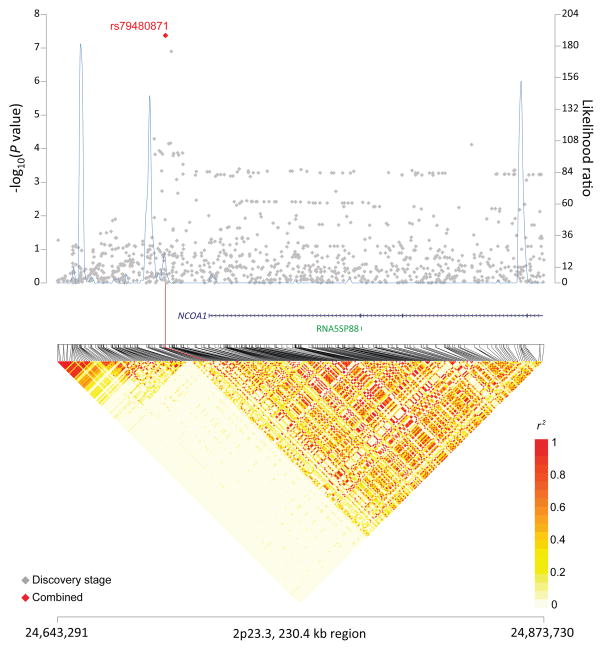
In a meta-analysis of all three stages (Supplementary Table 7), we identified four non-HLA SNPs in three novel loci at 6p25.3 (rs116446171, P=2.33×10−21) near EXOC2, 8q24.21 (rs13255292, P=9.98×10−13; rs4733601, P=3.63×10−11) near PVT1 and MYC, and 2p23.3 (rs79480871, P=4.23×10−8) near NCOA1 (Table 1; Figures 1a–c). The two 8q24.21 SNPs displayed minimal linkage disequilibrium (LD, r2=0.03 in 1000 Genomes CEU population). Furthermore, in conditional analysis, both rs13255292 (conditional OR=1.22, P =1.39×10−12) and rs4733601 (conditional OR=1.18, P =2.84×10−10) remained genome-wide significant; together these data support the presence of two independent SNPs associated with DLBCL at 8q24.21. We also observed two suggestive SNPs (P<5×10−7) (Supplementary Table 8), one at 5q31.3 (rs79464052, P=5.57×10−8) in ARAP3 (Supplementary Figure 4), and one at 3q13.33 (rs2681416), although the latter SNP did not replicate in stage 2 or 3.
Table 1.
Association of novel loci and new independent SNPs wth risk of diffuse large B-cell lymphoma (DLBCL)
| Location | Nearest gene(s) | SNP | Positiona | Risk alleleb | Other allele | RAFc | Stage | No. Cases/No. controls | OR | (95% CI) | P | Phet | I2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6p25.3 | EXOC2 | rs116446171 | 484,453 | G | C | 0.019 | Stage 1 | 2,661/6,220 | 2.26 | (1.82–2.81) | 1.48×10−13 | ||
| 0.018 | Stage 2 | 1,194/1,443 | 2.70 | (1.84–3.96) | 3.99×10−7 | ||||||||
| 0.019 | Stage 3 | 1,351/4,460 | 1.78 | (1.29–2.46) | 0.00040 | ||||||||
| Combined | 5,206/12,123 | 2.20 | (1.87–2.59) | 2.33×10−21 | 0.17 | 32.82 | |||||||
| 8q24.21 | PVT1 | rs13255292 | 129,076,573 | T | C | 0.321 | Stage 1 | 2,661/6,221 | 1.19 | (1.11–1.28) | 1.25×10−6 | ||
| 0.315 | Stage 2 | 1,195/1,444 | 1.30 | (1.14–1.47) | 4.29×10−5 | ||||||||
| 0.330 | Stage 3 | 1,322/4,498 | 1.22 | (1.09–1.36) | 0.001 | ||||||||
| Combined | 5,178/12,163 | 1.22 | (1.15–1.29) | 9.98×10−13 | 0.37 | 8.30 | |||||||
| rs4733601 | 129,269,466 | A | G | 0.477 | Stage 1 | 2,661/6,221 | 1.19 | (1.11–1.27) | 4.22×10−7 | ||||
| 0.479 | Stage 2 | 1,196/1,445 | 1.19 | (1.05–1.33) | 0.004 | ||||||||
| 0.487 | Stage 3 | 1,337/4,523 | 1.19 | (1.07–1.32) | 0.0016 | ||||||||
| Combined | 5,194/12,189 | 1.18 | (1.11–1.25) | 3.63×10−11 | 0.09 | 43.85 | |||||||
| 6p21.33 | HLA-B | rs2523607 | 31,322,790 | A | T | 0.120 | Stage 1 | 2,661/6,221 | 1.45 | (1.29–1.64) | 7.10×10−10 | ||
| 0.123 | Stage 2 | 1,195/1,444 | 1.14 | (0.96–1.35) | 0.14 | ||||||||
| 0.109 | Stage 3d | 1,114/1,102 | 1.25 | (1.04–1.51) | 0.019 | ||||||||
| Combined | 4,970/8767 | 1.32 | (1.21–1.44) | 2.40×10−10 | 0.26 | 21.63 | |||||||
| 2p23.3 | NCOA1 | rs79480871 | 24694472 | T | C | 0.076 | Stage 1 | 2,660/6,220 | 1.35 | (1.17–1.55) | 3.51×10−5 | ||
| 0.057 | Stage 2 | 1,195/1,443 | 1.56 | (1.22–1.99) | 0.00037 | ||||||||
| 0.063 | Stage 3 | 1,344/4,524 | 1.19 | (0.98–1.46) | 0.084 | ||||||||
| Combined | 5,199/12,187 | 1.34 | (1.21–1.49) | 4.23×10−8 | 0.15 | 34.59 |
Position according to human reference NCBI37/hg19;
Allele associated with an increased risk of DLBCL;
Risk allele frequency in controls;
Not genotyped in NCI Replication study.
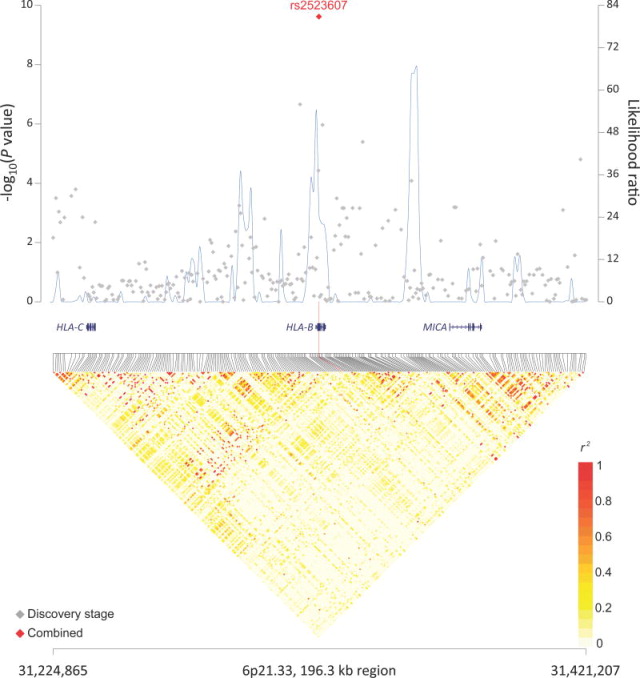
Figure 1. Association results, recombination hot-spots, and linkage disequilibrium (LD) plots for the regions newly associated with diffuse large B-cell lymphoma (DLBCL).
(a–d) Top, association results of GWAS data from stage 1 DLBCL-GWAS (grey diamonds) and combined data of stages 1–3 (red diamond) are shown in the top panels with −log10(P) values (left y axis). Overlaid are the likelihood ratio statistics (right y axis) to estimate putative recombination hotspots across the region on the basis of 5 unique sets of 100 randomly selected control samples. Bottom, LD heatmap based on r2values from combined control populations for all SNPs included in the GWAS. Shown are results for 6p25.3 (a), 8q24.21 (b), 2p23.36 (c), and p21.33 (d) regions.
Within the HLA region, rs2523607 (P=3.35×10−9) was carried forward for replication in stage 3. This SNP, localized at 6p21 in HLA-B, reached a combined P=2.40×10−10 in a meta-analysis of all three stages (Table 1; Figure 1d). To further evaluate the association of HLA variants with DLBCL risk, we imputed classical HLA alleles at six loci (HLA-A, B, C, DRB1, DQA1, and DQB1) in the four GWAS datasets from stages 1–2 and conducted a meta-analysis (Online Methods). The imputation accuracy of HLA types was high (>95.2%) when compared to HLA sequencing (four-digit resolution) previously performed on a subset of the NCI samples14 scanned as part of this study in stage 1 (Online Methods). Of all SNPs and classical HLA alleles tested across the MHC, only the SNP rs2523607 (OR=1.34, P=3.3×10−9 in stages 1 and 2) and the classical allele HLA-B*08:01 (OR=1.30, P=3.16×10−8 in stages 1 and 2) reached genome-wide significance (Supplementary Table 9). These markers were in very high LD (r2=0.91), and after adjusting for the effect of HLA-B*08:01, the association of rs2523607 was greatly weakened (P=5.5×10−3).
To gain additional insight into potential biological mechanisms, expression quantitative trait loci (eQTL) analyses were performed in two datasets consisting of lymphoblastoid cell lines (Online Methods). In one of the datasets, significant associations were observed for rs116446171 with HIST1H3F and rs2523607 with HCG27 (Supplementary Table 10), while in the other dataset significant associations (FDR<0.05) were observed for rs2523607 (using rs3130923 as a proxy, r2=0.94) with LY6G6E, FLOT1, and RNF5 (Supplementary Table 11); no associations were observed for the other DLBCL-associated loci.
To explore plausible mechanisms for the non-coding variants identified in our GWAS, the sentinel SNPs and those in high linkage disequilibrium (r2≥0.8) in Europeans in the 1000 Genomes Project were analyzed using HaploReg v215 (Online Methods; Supplementary Table 12). In addition, B-cell specific chromatin dynamics were assessed in a lymphoblastoid cell line (GM12878) using ChroMoS16, which utilizes the pre-computed chromatin state data for 9 cell lines (including GM12878)17. Of the 173 SNPs queried, 61 had information for GM12878 (Supplementary Figure 5), and 3 SNPs were identified as active or weak promoters only in GM12878, while 22 SNPs were identified as strong or weak enhancers in GM12878. In the other 8 cell lines, these regions were mostly defined as neutral, weakly transcribed or polycomb repressed. These results suggest that some of our SNPs are within regions of active chromatin state predominantly within B cells and have a role in the B-cell cis-regulatory network. These results are consistent with growing evidence that disease variants from GWAS are more likely to map to active chromatin sites than neutral sites, as was shown recently for systemic lupus erythematosus17. HaploReg showed that the majority of DLBCL-related SNPs were observed in regions of DNAse hypersensitivity common across multiple cell lines (e.g., rs116446171, rs2523607, rs13255292, rs4733601 near EXOC2, HLA-B, PVT1 or 7SK) whereas rs147193201 was specific to B-cells. The preponderance of DNAse hypersensitivity points to the existence of motifs, such as enhancers, silencers, promoters, insulators and other control elements of gene regulation. The proteins bound at these sites are known transcription factors such as NF-κB, c-MYC, GATA2 or genes that regulate transcription such as POL24H8, USF1 or POL2. These suggested mechanisms of action will require laboratory follow-up.
The susceptibility locus at 6p25.3 (rs116446171) maps near a plausible DLBCL candidate gene, EXOC2 (exocyst complex component 2), which is part of a large multiprotein complex responsible for vesicle trafficking and maintenance and intercellular transfer of viral proteins and virions18. EXOC2 functions at the interface between host defense and cell death regulation19. EXOC2 interacts with Ral proteins, and the Ral-exocyst regulatory node has a crucial role in the maintenance of epithelial cell polarity, cell motility and cytokinesis20,21, and in proliferation and metastasis20,22. It is notable that IRF4 is centromeric to EXOC2 and genetic variation in this region has been linked with chronic lymphocytic leukemia (CLL) risk23,24, and nominally to DLBCL risk25. However, rs116446171 was not in LD with the IRF4 CLL GWAS SNP rs87207123.
Two 8q24.21 variants (Figure 1b), rs13255292 and rs4736601 positioned at chr8:129.07Mb and chr8:129.26Mb, respectively, are approximately 1Mb telomeric to the 8q24 region linked with multiple cancers26, including CLL27. Both variants are in close proximity to PVT1, which is a non-coding RNA implicated in the MYC activation. Notably, a variant at 8q24.21 (rs2019960) has been linked to HL28, but the pair-wise r2 values of this SNP with both of our SNPs were low (r2<0.02). The close proximity of PVT1 and the MYC oncogene, which is known to be deregulated in Burkitt lymphoma29,30 and some DLBCLs 31,32, suggests that germline variation in this region could also contribute to DLBCL risk.
The susceptibility locus at 2p23.3 (rs79480871) maps near NCOA1, nuclear receptor coactivator 1 and ITSN2, intersectin 2. The former gene acts as a transcriptional coactivator for steroid and nuclear hormone receptors and is a member of the p160/steroid receptor coactivator (SRC) family33, while the latter gene encodes a protein that is a member of a family of proteins involved in clathrin-mediated endocytosis34 and may also augment the induction of T-cell receptor endocytosis35. However, our bioinformatics analysis did not identify a clear link to genes in this region, supporting the need to refine this signal in future work.
Through imputation with SNP2HLA,36 our strongest associations in the HLA region were with the HLA-B SNP rs2523607 and HLA-B*08:01, which are in very high LD, and based on our available sample size we cannot definitively rule out an orthogonal effect of rs2523607 in favor of HLA-B*08:01. HLA-B encodes the HLA class I heavy chain paralogue, which heterodimerizes with a light chain (β2 microglobulin) to play a central role in presenting intracellularly processed self or foreign antigens to CD8+ cytotoxic T lymphocytes. Class I molecules have been linked to a variety of immune-mediated diseases and cancers including HL, follicular lymphoma, DLBCL7,14,37,38, and more recently marginal zone lymphoma (Vijai, submitted). Our results strongly suggest HLA-B*08:01 as the primary MHC association with DLBCL risk. This classical allele is carried by the so-called ancestral 8.1 haplotype associated with other complex diseases (e.g., type I diabetes).39 Classical alleles of other HLA loci may also be involved (including those on the 8.1 haplotype), but larger sample sizes will be required to evaluate this question.
Our study represents the largest DLBCL GWAS in individuals of European descent. We did not observe a notable signal for a locus previously reported for DLBCL on 3q27 in East Asia10, rs6773854 (reported as OR=1.47, P=1.14×10−11), which was based on a discovery set of 253 B-cell NHL cases (148 DLBCLs). Although our current study had a similar MAF of 0.22 among controls, we observed an OR=1.06 and a P-value of 0.81 for this SNP (Supplementary Table 13), suggesting that the reported marker may not be correlated with the functional susceptibility allele in Europeans. Of the two suggestive loci (P<5×10−7) reported in the literature8,40, we did not observe an association for rs751837 with DLBCL (OR=0.97, P=0.46), identified in a small Japanese GWAS (OR=3.51, P=3.3×10−7)40, but we did observe a consistent albeit attenuated association for rs10484561 (OR=1.18, P=1.5×10−4) which was initially reported on a subset of the studies in stage 1 (OR=1.36, P=1.46×10−7)8. Previously, an InterLymph study of ~1,800 DLBCLs and ~6,500 controls reported a strong signal for a dinucleotide haplotype in the LTA/TNF locus (LTA 252A>G/TNF–308G>A) at 6p21.3 (OR=1.31, P=2.9 x10−8)5. Although nearly all of the cases from the previous publication were included in our current GWAS, the signal we observed overall was weaker (OR=1.15, P=8.5×10−4). The attenuation was not explained by study design (case-control, cohort) or adjustment for population substructure (data not shown), but could be due to population sampling differences, heterogeneity, or chance.
To explore the heritability of DLBCL, we estimated the contribution of all common SNPs to the variance explained by fitting all genotyped autosomal SNPs simultaneously using the method proposed by Yang et al41 in the Stage 1 dataset. We estimated that common SNPs, including but not limited to the loci discovered in this study, explain approximately 16% of the variance for DLBCL overall.
In summary, our findings represent an important step in defining the contribution of common genetic variants to risk for DLBCL. Our findings are notable because we have newly defined associations of several regions with susceptibility to DLBCL, and these regions harbor plausible candidate genes for further investigation. Further studies are required to discover additional common susceptibility loci as well as functional analyses that can explain the biological underpinnings of these new susceptibility loci.
ONLINE METHODS
Stage 1: DLBCL-GWAS
As part of a larger initiative, we conducted a genome-wide association study (GWAS) of diffuse large B-cell lymphoma (DLBCL) using cases and controls of European descent from 22 studies of non-Hodgkin lymphoma (NHL) (Supplementary Table 1), including nine prospective cohort studies, eight population-based case-control studies, and five clinic or hospital-based case-control studies. All studies were approved by their respective Institutional Review Boards, and informed consent was obtained for all participants. Cases were ascertained from cancer registries, clinics or hospitals, or through self-report verified by medical and pathology reports. To determine NHL subtype, phenotype data for all NHL cases were harmonized to the hierarchical classification proposed by the InterLymph Pathology Working Group42,43 based on the World Health Organization (WHO) classification44.
All DLBCL cases with sufficient DNA (n=2,878) and a subset of controls (n=2,854) frequency matched by age, sex, and study to the entire group of NHL cases, along with 4% quality control duplicates, were genotyped on the Illumina OmniExpress at the NCI Cancer Genomic Research Laboratory (CGR). Genotypes were called using Illumina GenomeStudio software, and quality control duplicates showed >99% concordance. Monomorphic SNPs and SNPs with a call rate of <95% were excluded. Samples with a call rate of ≤93%, mean heterozygosity <0.25 or >0.33 based on the autosomal SNPs, or gender discordance (>5% heterozygosity on X chromosome for males and <20% heterozygosity on the X chromosome for females) were excluded. Furthermore, unexpected duplicates (>99.9% concordance) and first-degree relatives based on identity by descent (IBD) sharing with Pi-hat>0.40 were excluded. Ancestry was assessed using the GLU struct.admix module based on the method by Pritchard et al.45 and participants with <80% European ancestry were excluded (Supplementary Figure 6). After exclusions, 2,661 (92.5%) cases and 2,685 (94.1%) controls remained (Supplementary Table 2). Genotype data previously generated on the Illumina Omni2.5 from an additional 3,536 controls from three of the studies (ATBC, CPSII, and PLCO) were also included11, resulting in a total of 2,661 cases and 6,221 controls for the stage 1 analysis. Of these additional 3,536 controls, 703 (~235 from each study) were selected to be representative of their cohort and cancer-free11, while the remainder were cancer-free controls from an unpublished study of prostate cancer in the PLCO. SNPs with call rate <95%, with Hardy-Weinberg equilibrium P-value<1×10−6, or with a minor allele frequency <1% were excluded from analysis, leaving 611,844 SNPs for analysis (Supplementary Table 4). To evaluate population substructure, a principal components analysis (PCA) was performed using the Genotyping Library and Utilities (GLU), version 1.0, struct.pca module, which is similar to EIGENSTRAT46. Plots of the first five principal components are shown in Supplementary Figure 7. Association testing was conducted assuming a log-additive genetic model, adjusting for age, sex, and four significant principal components. All data analysis and management was conducted using GLU.
Stage 2: In Silico Analysis of Three Independent DLBCL GWAS
Three independent DLBCL GWAS provided genotyping data for a meta-analysis, (Supplementary Table 1), which included data generated with the following commercial, SNP microarrays: Illumina HumanHap 660W for Mayo (393 DLBCL and 172 controls), HumanCNV370-Duo for UCSF7 (254 DLBCLs and 748 controls), and HumanHap 610K for GELA (549 cases). In all studies, subjects with a genotyping call rate <95%, duplicates, related individuals, and SNPs with a call rate <95% were removed prior to imputation (Supplementary Table 4). The GELA study was conducted on cases only; controls were drawn from a pool of 928 individuals from the French component of the EPIC cohort, who were previously scanned on Illumina HumanHap 660W or 610K47,48. We subsequently chose a subset of 525 individuals with matched ancestry as determined from the principal components analysis. In total, there were 1,196 cases and 1,445 controls in stage 2.
Imputation was conducted separately for each study in stages 1 and 2 using IMPUTE213 and the 1000 Genomes Project version 312. The imputation analysis was restricted to common SNPs (cut-off MAF>0.01 with imputation accuracy INFO score >0.3).
Association testing was conducted for each study using SNPTEST version 2, adjusting for age, sex, and any significant principal components. We evaluated the top 10 eigenvectors for the GELA, Mayo and UCSF studies, respectively, in each baseline risk model adjusting for both age and gender. Based on the significance level (P<0.05) of the regression coefficient for eigenvectors, we chose to adjust for three eigenvectors (EV1, EV7 and EV8) for GELA in the final association model, while no eigenvectors met criteria for adjustment of either the Mayo or UCSF studies.
All meta-analyses were performed using the fixed effects inverse variance method based on the beta estimates and standard errors from each study.
Stage 3: Replication studies and technical validation
In stage 3, eight SNPs in the most promising loci outside of the HLA region and one SNP from the HLA region (Supplementary Table 7) were taken forward for de novo replication in an additional 1359 cases and 4557 controls from four studies (Supplementary Table 1), except for rs2523607, which was not genotyped in one of the studies (NCI replication). Genotyping was conducted using custom TaqMan genotyping assays (Applied Biosystems) at the NCI Cancer Genomics Research Laboratory. Each assay was optimized and validated with 270 HapMap samples and additional CEPH samples (SNP500Cancer), and these samples were used as genotyping controls for clustering and reproducibility. All validated assays had 99% or higher concordance with HapMap and completion with control DNA was >97%. Blind duplicates from stage 3 samples (64 pairs; ~3%) yielded 100% concordance.
In technical validation, we observed a high correlation of genotyping calls from the OmniExpress microarray with confirmatory TaqMan assays in 455 stage 1 duplicate samples for two genotyped (rs13255292, r2=1.00; rs4733601, r2=1.00) and four imputed (rs116446171, r2=0.92; rs2523607, r2=0.99; rs2681416, r2=1.00; rs79480871, r2=0.94) SNPs. We also observed a high correlation of genotyping calls from the Illumina HumanHap 660W microarray with confirmatory TaqMan assays in stage 2 duplicate samples from the Mayo study (N=165) for two genotyped (rs13255292, r2=1.00; rs4733601, r2=1.00) and four imputed (rs116446171, r2=1.00; rs2523607, r2=1.00; rs79480871, r2=0.85; rs79464052, r2=0.95) SNPs.
HLA imputations and analysis
We imputed dense SNPs as well as classical HLA alleles (A, B, C, DRB1, DQA1, DQB1) and coding variants across the HLA region (chr6:29.5–33.2Mb, hg19) in the stage 1 (NCI) and stage 2 (MAYO, USCF2 and GELA/EPIC) studies using SNP2HLA36. The imputation was based on a reference panel from the Type 1 Diabetes Genetics Consortium (T1DGC), and consisted of genotypes from 5,225 individuals of European descent who were typed for HLA-A, B, C, DQA1, DQB1, DRB1, DPA1, DPB1 4 digit alleles. To assess imputation accuracy, we compared the imputed HLA alleles to HLA sequencing data (to 4 digits) available on a subset of samples from the NCI GWAS14, and found high concordance rates for HLA-A (97.3%), B (98.5%), C (98.1%) and DRB1 (97.5%). Due to the limited number of SNPs (N=7,253) in the T1DGC reference set, imputation of HLA SNPs was conducted with IMPUTE2 and the 1000 Genomes reference set as described above. A total of 68,488 SNPs, 201 classical HLA alleles (two- and four-digit resolution) and 1,038 AA markers including 103 AA positions that were ‘multi-allelic’ with three to six different residues present at each position, were successfully imputed (info score >0.3 for SNPs or r2>0.3 for alleles and AAs) and available for analysis. Multi-allelic markers were analyzed as binary markers (e.g., allele present or absent) and using a global test, and a meta-analysis was conducted where we tested SNPs, HLA alleles and AAs across the HLA region for association with DLBCL using PLINK49 or SNPTEST as described above.
Expression quantitative trait loci (eQTL) analysis
To evaluate the effect of our top loci (and SNPs in LD based on r2>0.8 in HapMap-CEU release 28) on gene expression, we conducted an eQTL analysis on lymphoblastoid cell lines using two independent datasets: Childhood asthma50 and HapMap51. For the childhood asthma dataset 50, peripheral blood lymphocytes were transformed into lymphoblastoid cell lines for 830 parents and offspring from 206 families of European ancestry. Using extracted RNA, gene expression was assessed with the Affymetrix HG-U133 Plus 2.0 chip. Genotyping was conducted using the Illumina Human1M Beadchip and Illumina HumanHap300K Beadchip, and imputation was performed using data from the 1kGP. All SNPs selected for replication were tested for cis associations (defined as gene transcripts within 1 Mb), assuming an additive genetic model, adjusting for non-genetic effects in the gene expression value. To gain insight into the relative importance of associations with our SNPs compared to other SNPs in the region, we also conducted conditional analyses, in which both the DLBCL SNP and the most significant SNP for the particular gene transcript (i.e., peak SNP) were included in the same model. Only cis associations that reached P<6.8×10−5, which corresponds to a false-discovery rate (FDR) of 1%, are reported (Supplementary Table 10).
The HapMap dataset consisted of a publicly available RNAseq dataset51 from transformed lymphoblastoid cell lines from 41 CEPH Utah residents with ancestry from northern and western Europe (HapMap-CEU), samples available from the Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE16921. Genotyping data for the same HapMap-CEU individuals were directly downloaded from HapMap (www.hapmap.org). Since rs2523607, rs79480871 and rs116446171 were not genotyped in HapMap, we selected rs3130923, rs6746301 and rs7762424 as respective proxies, as they were the strongest linked SNPs available in HapMap (r2=0.94, 0.69 and 0.54 in 1kGP-CEU, respectively). Correlation between expression and genotype for each SNP-probe pair was tested using the Spearman’s rank correlation test with t-distribution approximation and were estimated with respect to the minor allele in HapMap-CEU. P-values were adjusted using the Benjamini-Hochberg false-discovery rate (FDR) correction and eQTLs were considered significant at an FDR≤0.05 (Supplementary Table 11).
Bioinformatics: ENCODE and Chromatin State Dynamics
Using 1000 Genomes data, we identified SNPs with r2≥0.8 with our sentinel SNP that were reported to be non-synonymous or nonsense variants. We utilized HaploReg v215, which is a tool for exploring non-coding functional annotation using ENCODE data, to evaluate the genome surrounding our SNPs (Supplementary Table 12). To assess chromatin state dynamics, we used Chromos16, which has pre-computed data from ENCODE on 9 cell types based on Chip-Seq analyses17. These pre-computed data have genome-segmentation performed using multivariate hidden Markov-model to reduce the combinatorial space to a set of interpretable chromatin states. The output from Chromos separates data into 15 chromatin states corresponding to repressed, poised and active promoters, strong and weak enhancers, putative insulators, transcribed regions, and large-scale repressed and inactive domains (Supplementary Figure 5).
Heritability analyses
To estimate the contribution of all common SNPs to the variance explained, we used the method proposed by Yang et al41, which was extended to dichotomous traits52 and implemented in the Genome-wide Complex Trait Analysis (GCTA) software53. The genetic similarity matrix was estimated from our stage 1 data using all genotyped autosomal SNPs with a minor allele frequency >0.01. We used restricted maximum likelihood (REML), the default option for GCTA, to fit the appropriate variance components model that included the top 10 eigenvectors as covariates. The final estimate of heritability on the underlying liability scale assumed that the lifetime risk of DLBCL was 0.007454.
Estimate of recombination hotspots
To identify recombination hotspots in the region we used SequenceLDhot55, a program that uses the approximate marginal likelihood method56 and calculates likelihood ratio statistics at a set of possible hotspots. We tested five unique sets of 100 control samples. PHASE v2.1 program was used to calculate background recombination rates57,58 and LD heatmap was visualized in r2 using snp.plotter program59.
Supplementary Material
Acknowledgments
We thank C. Allmer, E. Angelucci, A. Bigelow, S. Buehler, K. Butterbach, A. Chabrier, J.M. Conners, M. Corines, M. Cornelis, K. Corsano, H. Dykes, L. Ershler, A. Gabbas, R.P. Gallagher, R.D. Gascoyne, P. Hui, L. Irish, L. Jacobus, L. Klareskog, A.S. Lai, J. Lunde, M. McAdams, R. Montalvan, L. Padyukov, M. Rais, T. Rattle, L. Rigacci, K. Snyder, G. Specchia, M. Stagner, G. Thomas, C. Tornow, G. Wood, and M. Yang.
The overall GWAS project was supported by the intramural program of the US National Institutes of Health/National Cancer Institute. A list of support provided to individual studies is provided in the Supplementary Note.
Footnotes
AUTHORS CONTRIBUTIONS
J.R.C., S.I.B., S.S.W., A.N., A.R.B.-W., Q.L., G. Severi, M. Melbye, L.R.T., M.P.P., C.L., B.M.B., S.L.S., S.d.S., K.E.S., C.F.S., N.R. and S.J.C. organized and designed the study. J.R.C., L.C., L.B., A.H., P.M.B., E.A.H., S.L.S., G. Salles, C.F.S., N.R. and S.J.C. conducted and supervised the genotyping of samples. J.R.C., S.I.B., V.J., Z.W., M.Y., L.C., P.I.W.d.B., D.C., J.G., D. Zhi, Y.W.A., J.H., B.M., J.S., L.L., J.P., C.C.C., N.C., S.d.S., K.E.S., C.F.S., N.R. and S.J.C. contributed to the design and execution of statistical analysis. J.R.C., S.I.B., V.J., H.G., J.M., S.S.W., Z.W., M.Y., L.C., A.N., D.C., A.M., C.R.F., A.J.D.R., C.L., K.E.S., C.F.S., N.R. and S.J.C. wrote the first draft of the manuscript. J.R.C., V.J., H.G., J.M., S.S.W., L.C., A.N., L.B., A.M., A.R.B.-W., Q.L., G. Severi, M. Melbye, J.G., R.D.J., E.K., L.R.T., M.P.P., C.M.V., J.J.S., G.G.G., D.A., R.S.K., M.Z., K.A.B., A.Z.-J., T.M.H., B.K.L., A.J.N., A.D., Y.W.A., M.L., C.A.T., S.M.A., T.E.W., G.J.W., A.S.V., D. Zelenika, H.T., C.H., T.J.M., H.H., B.G., H.-O.A., P.M.B., J.R., M.T.S., E.A.H., W.C., P.H., L.M.M., R.K.S., L.F.T., K.E.N., N.B., Y.B., P. Boffetta, P. Brennan, L.F., M. Maynadie, A. Staines, T.L., S.C., A. Smith, E. Roman, W.R.D., K.O., A.Z., R.J.K., D.J.V., T.Z., Y.Z., T.R.H., A.K., J.T., M.C.S., J.C., J. Virtamo, S.W., E. Riboli, P.V., R.K., D.T., R.C.H.V., H.B., A.T., E.A., S.D.L., M.R., B.M.B., F.L., E.G., P.K., Y.Y., B.C.H.C., D.D.W., N.C., J.F.F.J., S.L.S., X.W., S.d.S., K.E.S., G. Salles, C.F.S. and N.R. conducted the epidemiological studies and contributed samples to the GWAS and/or follow-up genotyping. All authors contributed to the writing of the manuscript.
COMPETING INTERESTS
The authors declare no competing financial interests
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Flowers CR, Sinha R, Vose JM. Improving outcomes for patients with diffuse large B-cell lymphoma. CA Cancer J Clin. 2010;60:393–408. doi: 10.3322/caac.20087. [DOI] [PubMed] [Google Scholar]
- 3.Wang SS, et al. Family history of hematopoietic malignancies and risk of non-Hodgkin lymphoma (NHL): a pooled analysis of 10,211 cases and 11,905 controls from the International Lymphoma Epidemiology Consortium (InterLymph) Blood. 2007;109:3479–88. doi: 10.1182/blood-2006-06-031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldin LR, Bjorkholm M, Kristinsson SY, Turesson I, Landgren O. Highly increased familial risks for specific lymphoma subtypes. Br J Haematol. 2009;146:91–4. doi: 10.1111/j.1365-2141.2009.07721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skibola CF, et al. Tumor necrosis factor (TNF) and lymphotoxin-alpha (LTA) polymorphisms and risk of non-Hodgkin lymphoma in the InterLymph Consortium. Am J Epidemiol. 2010;171:267–76. doi: 10.1093/aje/kwp383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skibola CF, et al. Genetic variants at 6p21.33 are associated with susceptibility to follicular lymphoma. Nat Genet. 2009;41:873–5. doi: 10.1038/ng.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conde L, et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat Genet. 2010;42:661–4. doi: 10.1038/ng.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smedby KE, et al. GWAS of follicular lymphoma reveals allelic heterogeneity at 6p21.32 and suggests shared genetic susceptibility with diffuse large B-cell lymphoma. PLoS Genet. 2011;7:e1001378. doi: 10.1371/journal.pgen.1001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vijai J, et al. Susceptibility Loci associated with specific and shared subtypes of lymphoid malignancies. PLoS genetics. 2013;9:e1003220. doi: 10.1371/journal.pgen.1003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan DE, et al. Genome-wide association study of B cell non-Hodgkin lymphoma identifies 3q27 as a susceptibility locus in the Chinese population. Nat Genet. 2013;45:804–7. doi: 10.1038/ng.2666. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, et al. Improved imputation of common and uncommon SNPs with a new reference set. Nat Genet. 2012;44:6–7. doi: 10.1038/ng.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang SS, et al. Human leukocyte antigen class I and II alleles in non-Hodgkin lymphoma etiology. Blood. 2010;115:4820–3. doi: 10.1182/blood-2010-01-266775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barenboim M, Manke T. ChroMoS: an integrated web tool for SNP classification, prioritization and functional interpretation. Bioinformatics. 2013;29:2197–8. doi: 10.1093/bioinformatics/btt356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–9. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukerji J, Olivieri KC, Misra V, Agopian KA, Gabuzda D. Proteomic analysis of HIV-1 Nef cellular binding partners reveals a role for exocyst complex proteins in mediating enhancement of intercellular nanotube formation. Retrovirology. 2012;9:33. doi: 10.1186/1742-4690-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mantovani A, Balkwill F. RalB signaling: a bridge between inflammation and cancer. Cell. 2006;127:42–4. doi: 10.1016/j.cell.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 20.Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–40. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- 21.Issaq SH, Lim KH, Counter CM. Sec5 and Exo84 foster oncogenic ras-mediated tumorigenesis. Mol Cancer Res. 2010;8:223–31. doi: 10.1158/1541-7786.MCR-09-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kashatus DF. Ral GTPases in tumorigenesis: emerging from the shadows. Exp Cell Res. 2013;319:2337–42. doi: 10.1016/j.yexcr.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Bernardo MC, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40:1204–10. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- 24.Berndt SI, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet. 2013;45:868–76. doi: 10.1038/ng.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang SS, et al. Common gene variants in the tumor necrosis factor (TNF) and TNF receptor superfamilies and NF-kB transcription factors and non-Hodgkin lymphoma risk. PLoS One. 2009;4:e5360. doi: 10.1371/journal.pone.0005360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wacholder S, Yeager M, Liao LM. Invited commentary: more surprises from a gene desert. Am J Epidemiol. 2012;175:488–91. doi: 10.1093/aje/kwr429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crowther-Swanepoel D, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010;42:132–6. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enciso-Mora V, et al. A genome-wide association study of Hodgkin’s lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3) Nat Genet. 2010;42:1126–30. doi: 10.1038/ng.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graham M, Adams JM. Chromosome 8 breakpoint far 3′ of the c-myc oncogene in a Burkitt’s lymphoma 2;8 variant translocation is equivalent to the murine pvt-1 locus. Embo J. 1986;5:2845–51. doi: 10.1002/j.1460-2075.1986.tb04578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love C, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–5. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savage KJ, et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114:3533–7. doi: 10.1182/blood-2009-05-220095. [DOI] [PubMed] [Google Scholar]
- 32.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–7. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onate SA, Tsai SY, Tsai MJ, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–7. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 34.Novokhatska O, et al. Adaptor proteins intersectin 1 and 2 bind similar proline-rich ligands but are differentially recognized by SH2 domain-containing proteins. PloS One. 2013;8:e70546. doi: 10.1371/journal.pone.0070546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGavin MK, et al. The intersectin 2 adaptor links Wiskott Aldrich Syndrome protein (WASp)-mediated actin polymerization to T cell antigen receptor endocytosis. J Exp Med. 2001;194:1777–87. doi: 10.1084/jem.194.12.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia X, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howell WM. HLA and disease: guilt by association. Int J Immunogenet. 2014;41:1–12. doi: 10.1111/iji.12088. [DOI] [PubMed] [Google Scholar]
- 38.Klitz W, Aldrich CL, Fildes N, Horning SJ, Begovich AB. Localization of predisposition to Hodgkin disease in the HLA class II region. Am J Hum Genet. 1994;54:497–505. [PMC free article] [PubMed] [Google Scholar]
- 39.Price P, et al. The genetic basis for the association of the 8.1 ancestral haplotype (A1, B8, DR3) with multiple immunopathological diseases. Immunol Rev. 1999;167:257–74. doi: 10.1111/j.1600-065x.1999.tb01398.x. [DOI] [PubMed] [Google Scholar]
- 40.Kumar V, et al. Common variants on 14q32 and 13q12 are associated with DLBCL susceptibility. J Hum Genet. 2011;56:436–9. doi: 10.1038/jhg.2011.35. [DOI] [PubMed] [Google Scholar]
- 41.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morton LM, et al. Proposed classification of lymphoid neoplasms for epidemiologic research from the Pathology Working Group of the International Lymphoma Epidemiology Consortium (InterLymph) Blood. 2007;110:695–708. doi: 10.1182/blood-2006-11-051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner JJ, et al. InterLymph hierarchical classification of lymphoid neoplasms for epidemiologic research based on the WHO classification (2008): update and future directions. Blood. 2010;116:e90–8. doi: 10.1182/blood-2010-06-289561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swerdlow S, Campo E, Harris N. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC Press; Lyon, France: 2008. [Google Scholar]
- 45.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 47.Schumacher FR, et al. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20:3867–75. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siddiq A, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21:5373–84. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 51.Cheung VG, et al. Polymorphic cis- and trans-regulation of human gene expression. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howlader N, et al. SEER Cancer Statistics Review, 1975–2010. National Cancer Institute; Bethesda, MD: Apr, 2013. http://seer.cancer.gov/csr/1975_2010/, based on November 2012 SEER data submission, posted to the SEER web site. [Google Scholar]
- 55.Fearnhead P. SequenceLDhot: detecting recombination hotspots. Bioinformatics. 2006;22:3061–6. doi: 10.1093/bioinformatics/btl540. [DOI] [PubMed] [Google Scholar]
- 56.Fearnhead P, Harding RM, Schneider JA, Myers S, Donnelly P. Application of coalescent methods to reveal fine-scale rate variation and recombination hotspots. Genetics. 2004;167:2067–81. doi: 10.1534/genetics.103.021584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165:2213–33. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crawford DC, et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat Genet. 2004;36:700–6. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- 59.Luna A, Nicodemus KK. snp.plotter: an R-based SNP/haplotype association and linkage disequilibrium plotting package. Bioinformatics. 2007;23:774–6. doi: 10.1093/bioinformatics/btl657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.