Figure 1.
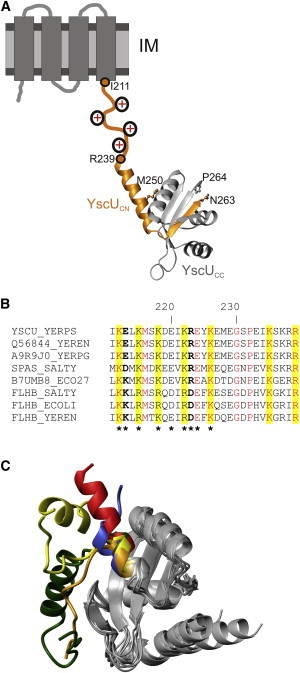
YscUC structure and domain arrangements. (A) Representation of the current structural biology view of YscU (25). The membrane-spanning domain (NTD) is illustrated on an IM as four cylinders, and the evolutionarily conserved linker sequence spanning residues Ile211 and Gln239 is illustrated as an orange line with conserved positive charges added schematically. YscUC is represented by the crystallographic structure (Protein Data Bank (PDB) code 2JLI (23)). The polypeptides resulting from autoproteolytic cleavage at the (N↑PTH) motif are indicated in orange (YscUCN) and gray (YscUCC). The C-terminal residue in YscUCN (Asn263) and the N-terminal residue in YscUCC (Pro264) are indicated. Met250, the first structured residue of YscUC in solution is highlighted. (B) The YscU linker sequence contains conserved positively charged side chains. Multiple sequence alignment of YscU (Y. pseudotuberculosis), SpaS (Salmonella typhimurium), EscU (Escherichia coli), and FlhB families from different organisms. Conserved residues that are positively charged are highlighted in yellow. In YscU and SpaS sequences, residues at positions 213 and 223 (bold) are negatively or positively charged, whereas they are oppositely charged in the different FlhB proteins. Asterisks indicate residues that were mutated in this study. (C) Structural plasticity of the N-terminus in YscUC and orthologs. Crystallographic structures are superimposed on the core of the autoproteolytic domains using the DALI server (26). The proteins can be identified by the colors of their N-terminal segments: red, Y. pestis YscUC (PDB 2JLI (23)); green, Y. enterocolitica YscUCN263A (PDB 2V5G (25)); yellow, E. coli EscUC (PDB 3BZL (22)); orange, E. coli EscUCT264A (PDB 3BZV (22)); and blue, S. typhimurium SpaSC (PDB 3C01 (22)).
