Figure 6.
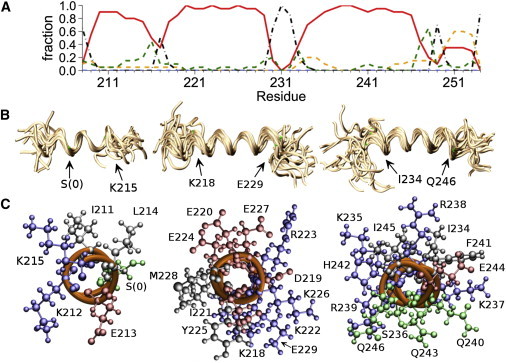
Structure of YscUCN in complex with SDS micelles computed from NMR restraints. (A) Occupation of different secondary-structure conformations by each residue, averaged over the structure ensemble. Red, α-helix; dashed yellow, 310 helix; dashed green, turn; dash-dotted black, bend or coil. (B) Backbone alignments for helices 1–3, showing an ensemble of 20 low-energy conformers. Backbones are rendered as golden coils. Positions of Cα atoms at the edges of the aligned segments corresponding to the helix boundaries are marked with green dots and labeled with arrows. (C) View along the long axis (from C to N terminus) of the first three helices of YscUCN, emphasizing amphipathic helical residue distributions. Backbone atoms are rendered as orange coils and side chains in ball-and-stick format colored according to residue polarity and charge: blue, basic (H, K, and R); red, acidic (D and E); green, polar (S and Q); black, = nonpolar (I, F, M, and Y). Molecular structures were aligned and rendered with the VMD program (51).
