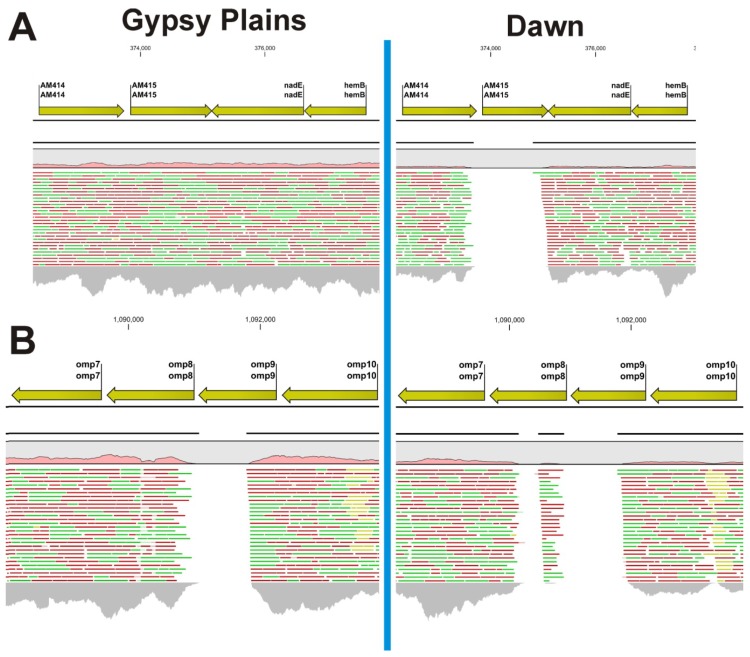
Figure 2.
Read mapping highlights large INDELs in the Gypsy Plains and Dawn strains. Mapping of 454 reads to the St. Maries genome of A. marginale highlights large INDELs in the Gypsy Plains and Dawn strains. Positions in the St. Maries genome are shown on top of the figure. Genes and their annotation in the St. Maries genome are shown with yellow arrows. A pink histogram shows coverage relative to whole genome mapping. Red, green and yellow lines represent the reads mapped to that particular region of the genome. Green reads have been mapped in forward direction; red reads have been mapped in reverse direction. Yellow reads are reads that have multiple equally good alignments to the reference. The CLC4 and GxWb5.5 mapping algorithms were used with the following parameters: mismatch cost 2, insertion cost: 3, deletion cost: 3, length fraction: 50%, and similarity fraction: 80%. The grey histogram at the bottom of the figure shows coverage depth for that particular region of the genome. (A) Shows the INDEL found in the Dawn strain for gene AM415. This 1193 bp long INDEL is only found in the non-transmissible/low virulence Dawn strain. Gene AM415 is present in the Gypsy Plains strain; (B) Shows the INDELs found at positions 1091010–1091800 in the Gypsy Plains strain. This INDEL affects the omp8 gene and is 791 bp long. Two INDELs were found for the Dawn strain in the same region of the genome. These two INDELS are found at positions 1090143–1090469 and 1090890–1091782, with 327 and 892 bp in length, respectively for the Dawn strain. While the INDEL for the AM415 gene was confirmed through PCR, amplification of the omp6-10 operon revealed a 1.5 kb deletion common to both Australian strains. The mapping discrepancies shown in this figure are likely due to sequence diversity of the Australian omp genes.