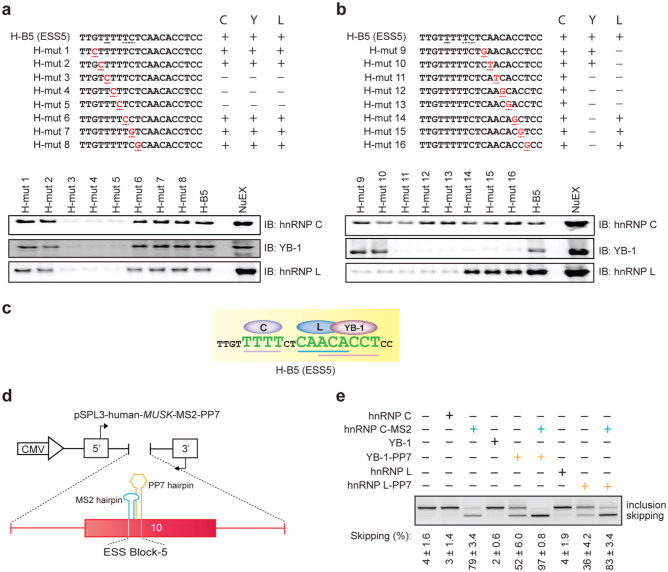
Figure 5. Binding of hnRNP C, YB-1, and hnRNP L to specific motifs enhances coordinated skipping of MUSK exon 10.
(a, b) Scanning mutagenesis to map the binding motifs of hnRNP C, YB-1, and hnRNP L in ESS5 (H-B5). RNA probe sequences are shown in the upper panels, where discordant nucleotides between human and mouse are underlined in H-B5 (ESS5). Artificial mutations are shown in red. RNA affinity-purified products are detected by immunoblotting in the lower panels. The results are indicated on the right side by “+” and “−” for positive and negative binding to each probe, respectively. (c) The resulting binding site of each factor from panels (a) and (b) is schematically shown. Essential binding nucleotides are indicated by large green letters and are underlined. (d) Schematic of a reporter minigene (pSPL3-human-MUSK-MS2-PP7). MS2 coat protein-binding hairpin RNA (blue) is substituted for the first half of ESS5 (binding site of hnRNP C). Similarly, PP7 coat protein-binding hairpin RNA (orange) is substituted for the second half of ESS5 (binding sites of hnRNP L and YB-1). (e) RT-PCR of pSPL3-human-MUSK-MS2-PP7 minigene in HeLa cells that are co-transfected with the indicated effectors. Blue and orange letters match to those in (d). The mean and SD (n = 3) of the ratio of exon skipping in each treatment is shown below the gel image.