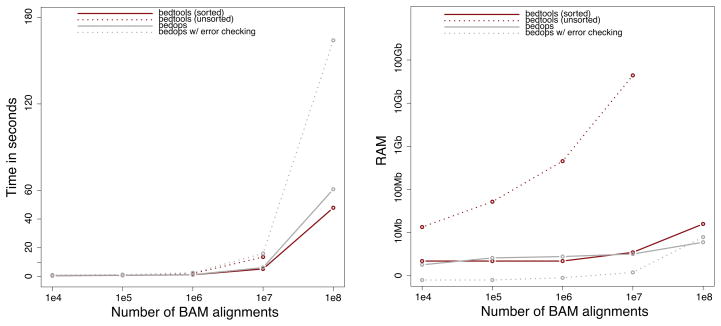
Figure 2. BEDTools scalability.
The runtime in seconds (left panel) and memory usage (right panel) are compared when using either unsorted genome intervals (dashed red) or genome intervals that have been pre-sorted in genome order (solid red). As a basis of comparison, the BEDTools performance is compared to the BEDOPS toolset, both with (solid gray) and without (dashed gray) automatic error checking.