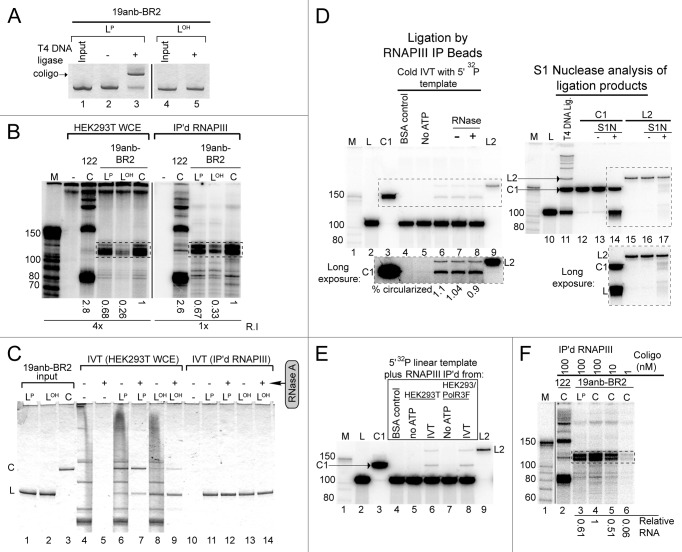
Figure 5. Coligo linear precursors nicked in a helical region are transcribed like intact coligos. (A) T4 DNA ligase assay to verify dephosphorylation of the 19anb-BR2 linear precursor. LOH, dephosphorylated linear precursor; LP, 5′ phosphorylated linear precursor. DNA visualized by Stains-All after DPAGE. (B) IVT of 100 nM 19anb-BR2 templates (LP, LOH, C) using HEK293T WCE or IP’d RNAP III. Coligo 122 was used as a positive IVT control. The relative amounts of monomer transcripts (boxed) were calculated separately for IP’d RNAP III and HEK293T WCE and normalized within each gel to coligo templated transcripts. R.I., relative exposure intensity of image. (C) Input templates were recovered from HEK293T WCE IVT reactions and visualized by Stains-All after DPAGE. RNase A was used to clarify results by degrading endogenous RNA. (D) About 1% of LP19anb-BR2 is circularized by IP’d RNAP III beads. 5′ 32P labeled 19anb-BR2 linear precursor DNA was examined at the end of an IP’d RNAP III IVT reaction (lanes 1–9). C1 is a gel mobility standard for coligo 19anb-BR2; L2 is a linear ligation dimer. BSA negative control substituted BSA protein for WCE during IP. No ATP lane is a negative control for DNA Ligase activity on the IP’d RNAP III beads. S1 Nuclease treatment (S1N) of C1 and L2 isolated from a T4 DNA ligase reaction on 5′-32P labeled linear precursor was used to check for circularization (lanes 10–17). Insets are longer exposures of boxed gel regions. (E) Control IP from unmodified HEK293T cell WCE also contained a contaminating DNA Ligase activity, indicating that the contaminating ligase activity is not RNAP III -dependent. (F) IP’d RNAP III IVT as a function of coligo 19anb-BR2 concentration. The 1 nM lane (lane 6, 0.06 relative RNA produced) represents the amount of circularized linear precursor formed by contaminating DNA ligase activity during IP’d RNAP III IVT. It cannot account for the amount of transcript formed by 100 nM linear template (lane 3, 0.61 relative RNA).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.