Figure.
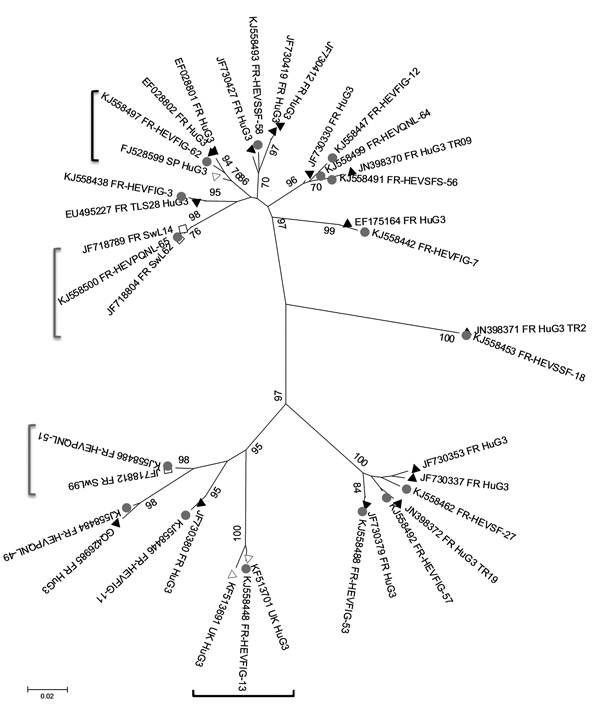
Phylogenetic tree of hepatitis E virus (HEV) sequences identified in food samples, France, 2011. Phylogenetic tree including 16 HEV sequences detected in food samples (gray circles) and the closest human (black triangles, French origin; white triangles, British or Spanish origin) or swine (white squares) sequences was constructed by using the neighbor-joining method with a bootstrap of 1,000 replicates based on the ClustalW alignment (MEGA4, http://www.megasoftware.net) on 290 nt sequences from open reading frame 2. HEV sequences retrieved from GenBank with >98% nt identity are indicated with their accession numbers. Bootstrap values of >70% are indicated on respective branches. Scale bar indicates nucleotide substitutions per site. Similar human and food sequences are shown in black brackets, similar swine and food sequences are shown in gray brackets.
