Table 1. Summary of performance for various fine-mapping methods.
| Variant Ranking Across All Loci | Variant Ranking Independently at Each Locus | ||||||
| Method | Summary Data | Incorporates Annotation | Assumed Num. of Causal Variants | Causals Identified | Causals Identified | ||
| 50% | 90% | 50% | 90% | ||||
| P-value | Yes | No | n/a | 5.74 | 12.60 | 2.94 | 19.15 |
| CAT score [12] | No | No | n/a | 2.04 | 13.29 | 2.56 | 17.80 |
| LLARRMA [13] | No | No | Mult | 1.98 | 21.93 | 2.46 | 23.11 |
| piMass-RB [14] | No | No | Mult | 2.83 | 16.31 | 2.18 | 15.15 |
| Maller et al. [5] | Yes | No | Single | 2.68 | 25.44 | 2.96 | 19.13 |
| fgwas (no annot) | Yes | No | Single | 2.69 | 25.48 | 2.95 | 19.11 |
| PAINTOR (no annot,1CV) | Yes | No | Single | 2.69 | 22.49 | 2.95 | 19.09 |
| fgwas [10] | Yes | Yes | Single | 1.95 | 24.77 | 2.05 | 17.37 |
| PAINTOR (1CV) | Yes | Yes | Single | 1.95 | 21.51 | 2.03 | 17.43 |
| PAINTOR (no annot) | Yes | No | Mult | 1.76 | 12.25 | 2.24 | 16.86 |
| PAINTOR | Yes | Yes | Mult | 1.26 | 10.42 | 1.61 | 13.68 |
| PAINTOR True | Yes | Yes | Mult | 1.23 | 10.22 | 1.59 | 13.48 |
Methods were benchmarked using the average number of SNPs per locus selected to find (50%,90%) of all causal variants. We simulated a trait with 100 risk loci explaining 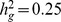 fine-mapped through sequencing of N = 10,000 samples and assessed accuracy only at loci that harbor at least one casual variant (64 loci on the average). We explored two methods to prioritizing variants: (1) “Variant Ranking Across All Loci” prioritizes SNPs across all loci while (2) “Variant Ranking Independently at Each Locus”, first prioritizes variants at each risk locus followed by merging across all loci. We note that PAINTOR 1CV and/or no annot corresponds to running PAINTOR assuming a single causal variant and/or not providing access to annotations. PAINTOR True did not empirically estimate enrichment but used the true enrichment values for each functional annotation data.
fine-mapped through sequencing of N = 10,000 samples and assessed accuracy only at loci that harbor at least one casual variant (64 loci on the average). We explored two methods to prioritizing variants: (1) “Variant Ranking Across All Loci” prioritizes SNPs across all loci while (2) “Variant Ranking Independently at Each Locus”, first prioritizes variants at each risk locus followed by merging across all loci. We note that PAINTOR 1CV and/or no annot corresponds to running PAINTOR assuming a single causal variant and/or not providing access to annotations. PAINTOR True did not empirically estimate enrichment but used the true enrichment values for each functional annotation data.
