Table 2. Leveraging functional priors leads to improved fine-mapping resolution.
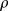 -level -level |
Method | Annotations | Causals Identified | SNPs Selected |
| 90% | Maller et al. | − | 64.2 | 265.0 |
| fgwas | + | 64.5 | 209.6 | |
| PAINTOR | − | 91.9 | 510.3 | |
| PAINTOR | + | 91.2 | 393.7 | |
| 95% | Maller et al. | − | 69.6 | 343.7 |
| fgwas | + | 70.2 | 290.8 | |
| PAINTOR | − | 97.2 | 687.8 | |
| PAINTOR | + | 97.0 | 567.2 | |
| 99% | Maller et al. | − | 77.7 | 506.6 |
| fgwas | + | 77.9 | 457.3 | |
| PAINTOR | − | 102.6 | 1074.4 | |
| PAINTOR | + | 102.7 | 954.3 |
We define an ρ-level confidence set as the number of SNPs we need to select in order to consume an 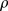 fraction of the total posterior probability mass over all loci. Results in the table correspond to averaging over 500 independent simulations where the average number of true causals SNPs per simulation was 109.2. The size of 90%, 95%, and 99% confidence sets are reduced by 22.8%, 17.5% and 11.1% when incorporating functional annotations as prior probabilities. Methods that assume one causal variant are miss-calibrated due to loci with multiple causals.
fraction of the total posterior probability mass over all loci. Results in the table correspond to averaging over 500 independent simulations where the average number of true causals SNPs per simulation was 109.2. The size of 90%, 95%, and 99% confidence sets are reduced by 22.8%, 17.5% and 11.1% when incorporating functional annotations as prior probabilities. Methods that assume one causal variant are miss-calibrated due to loci with multiple causals.
