A directed differentiation protocol for human embryonic stem cells (hESCs) that mimics the natural development process toward hyaline cartilage to generate chondrogenic cells with high efficiency and purity was previously reported. This study takes the work forward, optimizing the protocol, extending it to human induced pluripotent stem cells and demonstrating that hESC-derived chondrogenic cells can regenerate hyaline cartilage using an osteochondral defect model in nude rats.
Keywords: Cell transplantation, Embryonic stem cell, Tissue regeneration, Arthritis
Abstract
In initial work, we developed a 14-day culture protocol under potential GMP, chemically defined conditions to generate chondroprogenitors from human embryonic stem cells (hESCs). The present study was undertaken to investigate the cartilage repair capacity of these cells. The chondrogenic protocol was optimized and validated with gene expression profiling. The protocol was also applied successfully to two lines of induced pluripotent stem cells (iPSCs). Chondrogenic cells derived from hESCs were encapsulated in fibrin gel and implanted in osteochondral defects in the patella groove of nude rats, and cartilage repair was evaluated by histomorphology and immunocytochemistry. Genes associated with chondrogenesis were upregulated during the protocol, and pluripotency-related genes were downregulated. Aggregation of chondrogenic cells was accompanied by high expression of SOX9 and strong staining with Safranin O. Culture with PluriSln1 was lethal for hESCs but was tolerated by hESC chondrogenic cells, and no OCT4-positive cells were detected in hESC chondrogenic cells. iPSCs were also shown to generate chondroprogenitors in this protocol. Repaired tissue in the defect area implanted with hESC-derived chondrogenic cells was stained for collagen II with little collagen I, but negligible collagen II was observed in the fibrin-only controls. Viable human cells were detected in the repair tissue at 12 weeks. The results show that chondrogenic cells derived from hESCs, using a chemically defined culture system, when implanted in focal defects were able to promote cartilage repair. This is a first step in evaluating these cells for clinical application for the treatment of cartilage lesions.
Introduction
Cartilage forms the important load-bearing surface of articular joints, containing an expanded extracellular matrix maintained by a sparse population of chondrocytes. Because of its avascular nature, articular cartilage exhibits poor intrinsic capacity for repair and following trauma is predisposed to develop progressive osteoarthritis. An important aim is thus to repair focal defects and eventually larger osteoarthritic (OA) lesions with strategies to develop new cartilage tissue. This is very challenging because of the lack of suitable cell sources. Cell-based treatments using primary human chondrocytes and adult stem cells have delivered some success, but without evidence that they can provide a permanent or large-scale solution. Although autologous chondrocyte implantation has been applied to treat focal cartilage defects [1, 2] the invasive procedure and dedifferentiation of cultured chondrocytes during monolayer expansion hinders its wider application [3]. Furthermore, complications such as hypertrophy, resulting in vascular invasion and calcification, have impeded progress [4, 5]. Mesenchymal stem cells have the potential to form chondrocytes [6] and have been used for cartilage repair [7] but have limited capacity for proliferation and differentiation [8]. Human pluripotent stem cell-derived chondrocytes offer an alternative cell source for cell-based cartilage repair and may provide a consistent chondrocyte source to screen for novel compounds that delay cartilage degeneration. Induced pluripotent stem cells (iPSCs) will also provide the opportunity to generate joint disease models.
We previously reported a directed differentiation protocol for human embryonic stem cells (hESCs) that mimics the natural development process toward hyaline cartilage to generate chondrogenic cells with high efficiency and purity [9]. This protocol exploits the feeder-free and serum-free culture system we developed to provide potential GMP conditions for the growth and expansion of hESCs [10]. It has the potential for easy clinical adaptation because we are also generating new clinical grade hESC lines. To take the work forward, we optimized the protocol, extending it to human iPSCs, and demonstrate that hESC-derived chondrogenic cells can regenerate hyaline cartilage using an osteochondral defect model in nude rats.
Materials and Methods
Cell Culture and Directed Chondrogenic Differentiation
HUES1 and MAN7 cells were cultured as previously described [9]. Briefly, hESCs were cultured on mitomycin C-inactivated mouse embryonic fibroblasts (iMEFs) in hESC-medium [10]. For feeder-free culture, cells were lifted from the iMEF layers with TrypLE and plated onto fibronectin (FN)-coated (Millipore) tissue culture flasks with feeder-free medium [10]. The hESCs were differentiated with sequential addition of growth factors in the time sequence and at the concentrations previously described [9]. Briefly, Wnt3a, Activin-A, and BMP4 were applied to differentiate the hESCs toward primitive streak mesendoderm (days 1–3), followed by BMP4, Follistatin, and GDF5 to drive differentiation to mesoderm (days 4–8); finally, GDF5, FGF2, and NT4 were used to promote chondrogenesis (days 9–14). Cells were passaged three times in the previously published protocol (days 5, 8, and 12) with a change of coating substrate (FN, gelatin, or a mixture of both). To improve the protocol, we compared the effect of omitting the passage on day 12 or using a mixture of FN and gelatin instead of gelatin alone on the yield of chondrogenic cells (supplemental online Fig. 1).
Generation of iPSCs
Human dermal fibroblasts derived from the dermis of normal human adult skin were obtained from the European Collection of Animal Cell Cultures and were infected with a monocistronic human lentiviral noninducible reprogramming system, including OCT4 (iL O-9), SOX2 (iL S-9), KLF4 (iL K-9), and c-Myc (iL M-9) from Vectalys (Toulouse, France, http://www.vectalys.com). After transduction for 24 hours, the medium was replaced with complete Dulbecco’s modified Eagle’s medium (DMEM). The infected fibroblasts were harvested 6 days after transduction and replated on iMEFs in complete DMEM, which was replaced after 36 hours with hESC culture medium. Twenty-five to thirty days after transduction, ESC-like colonies were manually dissected for expansion (supplemental online Fig. 2A). iPSCs were characterized by expression of pluripotency markers, multilineage differentiation through embryoid body generation (supplemental online Fig. 2B) and production of teratomas (supplemental online Fig. 2C).
Generation of Enhanced Green Fluorescent Protein-Labeled hESCs
To label the ESCs with enhanced green fluorescent protein (EGFP), HUES1 cells were infected with lentiviral particles carrying elongation factor 1α (EF1α)-EGFP (pLVTHM, Addgene [Cambridge, MA, https://www.addgene.org] plasmid 12247). Four days after transduction, EGFP-positive cells were isolated by fluorescence-activated cell sorting and plated on iMEFs for expansion.
Gene Expression Analysis
Total RNA from cell cultures was extracted using mirVana miRNA isolation kit (Life Technologies, Rockville, MD, http://www.lifetech.com). Total RNA from human OA cartilage was a gift from Dr. Sara Dunn (University of Manchester) and was extracted using TRIzol. Genomic DNA was removed using Ambion DNase I (Life Technologies). Total RNA was reverse transcribed using Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI, http://www.promega.com). Expression of candidate genes was determined using SYBR Green PCR Master Mix (Applied Biosystems) with an ABI PRISM 7500 Real Time System (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com) using gene-specific oligonucleotide primers (supplemental online Table 1). Expression levels were normalized to GAPDH and calculated using the 2−ΔCt method. At least three independent differentiation experiments were performed.
Immunofluorescence
Cells cultured on fibronectin-coated plastic were fixed using 4% paraformaldehyde, blocked with 10% serum of the same species as the secondary antibody, incubated at 4°C for 16 hours with primary antibody (5 μg/ml SOX9; Millipore, Billerica, MA, http://www.millipore.com) or irrelevant antibody followed by 1 hour in appropriate secondary antibody in blocking buffer. For paraffin-embedded tissue sections, samples were deparaffinized, rehydrated, and subjected to antigen retrieval using sodium citrate buffer (0.01 M, pH 6). Then human vimentin (4 μg/ml; R&D Systems Inc., Minneapolis, MN, http://www.rndsystems.com) was stained using a similar method for detection of SOX9.
Flow Cytometry
Single cell suspensions were fixed in ice-cold methanol (10 minutes at −20°C) and permeabilized by incubation with 1% bovine serum albumin (BSA), 0.5% Triton X-100 in phosphate-buffered saline (PBS) for 15 minutes. Cells were incubated with primary antibody (mouse anti-OCT4, 1 μg/ml; goat anti-human SOX-9, 8 μg/ml) diluted in ice-cold blocking buffer (1% BSA in PBS) overnight at 4°C followed by appropriate secondary antibodies.
Osteochondral Defect Model and Histology/Immunohistochemistry
Twenty athymic RNU rats (Charles River Laboratories, Wilmington, MA, http://www.criver.com) weighing 200–250 g were used following local ethical committee approval and under U.K. Home Office license 40/4237. Osteochondral defects (2-mm diameter and approximately 2-mm depth) were created in the trochlear grooves of the distal femurs using a biopsy punch. Fibrin constructs were formed by resuspending 3 × 106 cells in 50 μl of fibrinogen followed by adding 50 μl of thrombin; under these conditions, the fibrin gel formed within seconds. Implants (containing 2 × 105 cells) were cut to fit the osteochondral defects using a biopsy punch for creating the defects (supplemental online Fig. 3A). Defects implanted with only fibrin gel served as controls. For histological analysis, specimens were fixed in 10% neutral buffered formalin overnight and, decalcified in 10% ethylenediaminetetraacetic acid (pH 7.4) for 2 weeks, and then embedded into paraffin. Sections were cut at 5 μm, deparaffinized, and stained with hematoxylin and eosin (HE), and immunohistochemistry (anti-Col2A1, 2 μg/ml, Santa Cruz; anti-Collagen I, 2 μg/ml; Abcam, Cambridge, U.K., http://www.abcam.com) was performed using diaminobenzidine histochemistry kits (Molecular Probes). For sulfated glycosaminoglycan (sGAG) detection, fixed cells were stained in 0.1% (wt/vol) Safranin O (in 0.1% [vol/vol] in acetic acid) for 5 minutes at room temperature. The specificity of Safranin O for sGAG was determined by incubating cultures with 1 unit/ml chondroitinase ABC (Sigma) in chondroitinase buffer (50 mM Tris [pH 8.0], 60 mM sodium acetate, 0.02% [wt/vol] BSA) for 30 minutes at 37°C before Safranin O staining. Some joints were also fixed and embedded in OCT for cryosectioning and used to detect the EGFP-labeled cells in the defect repair area. Histological scoring for cartilage repair was performed using Pineda’s method [11, 12].
Statistics
Data are shown as the means ± SD. An unpaired t test for data from two groups or one-way analysis of variance for data from groups of three or more was used to compare data. p values <.05 were considered to be significant.
Results
Optimization of the Directed Chondrogenic Differentiation Protocol
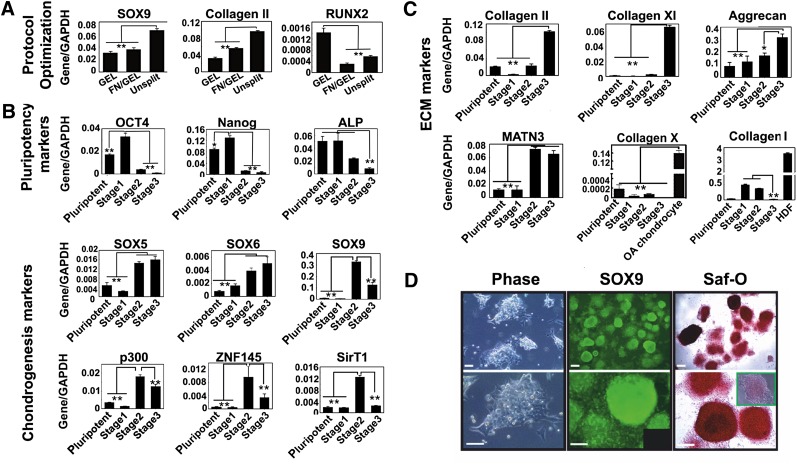
We optimized the published protocol to enhance chondrogenesis by avoiding splitting the cells on day 12 and keeping the cells on a 1:1 mixture of fibronectin and gelatin (supplemental online Fig. 1), and this promoted the expression of chondrogenic genes, including Sry-type HMG box 9 (SOX9) and COL2A1, a downstream target of SOX9 [13–15], which encodes the most abundant cartilage matrix protein collagen II (Fig. 1A). This improvement may result from maintaining a high cell density and accumulation of extracellular matrix (ECM), which provided an improved niche for chondrogenesis.
Figure 1.
Characterization of cells subjected to the directed differentiation protocol. (A): The directed differentiation protocol was optimized by avoiding splitting the cells on day 12 and continuation of fibronectin substrate with gelatin to the end of the protocol, which resulted in a higher expression level of SOX9 and COL2A1, but lower RUNX2. The results are the means ± SD (n = 3). (B, C): Expression of genes associated with pluripotency and chondrogenesis (B) and chondrocyte ECM (C) during the directed the differentiation protocol was analyzed by quantitative reverse transcription polymerase chain reaction. Human dermal fibroblasts were used as a positive control for Collagen I. The results are the means ± SD (n = 3). Levels of significance are depicted as follows: ∗, p < .05; ∗∗, p < .01. (D): Chondrogenic cells derived from MAN7 showed a very high level of aggregation at the end of the protocol and high expression of SOX9 (inset shows IgG control), as well as a high level of Safranin O staining (inset shows pretreated with chondroitinase ABC). Scale bars = 100 μm. Abbreviations: ALP, alkaline phosphatase; ECM, extracellular matrix; FN, fibronectin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GEL, gelatin; Saf-O, Safranin O.
Characterization of Gene Expression of hESCs During Chondrogenic Differentiation
Assessment of pluripotency-associated genes showed that OCT4 and Nanog expression were high in hESCs but were abolished by the end of the protocol and alkaline phosphatase (ALP) expression also decreased reflecting loss of pluripotency [16] (Fig. 1B). The expression of SOX9 correlated with zinc-finger protein 145 (ZNF145), an upstream regulator of SOX9 [17] that reached a peak at stage 2 before declining slightly at stage 3. However, the transactivation of SOX9 at stage 3 may be stabilized by high expression of SOX5 and SOX6, which act cooperatively with SOX9 to regulate the expression of COL2A1 and other genes of chondrogenic differentiation [18, 19] (Fig. 1B). P300, a histone acetyltransferase, modulates the function of SOX9 and other transcription factors [20] and was expressed at a consistently high level during differentiation. Interestingly, SIRT1, an NAD-dependent histone deacetylase, proposed to be a positive regulator of SOX9 [21], increased in parallel with SOX9 and also decreased at stage 3. Chondrocyte-ECM genes, collagen II and XI, showed a dramatic increase during differentiation with some increase in aggrecan (Fig. 1C), whereas collagen I expression was negligible. Matrilin-1 (MATN1) expression decreased at stage 3, whereas the expression of Matrilin-3 (MATN3) increased throughout and was around 50 times higher than that of MATN1 at the end of the protocol. This higher expression of MATN3 is associated with articular and not epiphyseal chondrocytes [22]. The expression of collagen X (COLX), a gene associated with chondrocyte hypertrophy [23], was negligible at all stages compared with the high expression detected in a sample of OA cartilage (800 times higher). The expression ratio of MATN3/MATN1, together with the negligible expression of COLX, suggests that the chondrogenic cells generated are on a developmental pathway to articular chondrocytes and not to epiphyseal chondrocytes. The revised protocol developed with HUES1 was also validated with a new hESC line, MAN7, which produced prominent SOX9 and Safranin O positive cell aggregates at the end of the protocol (Fig. 1D). The Safranin O intensity indicated chondroitin sulfate accumulation, which was confirmed by chondroitinase ABC digestion.
Absence of Pluripotent Cells in hESC Chondrogenic Cultures
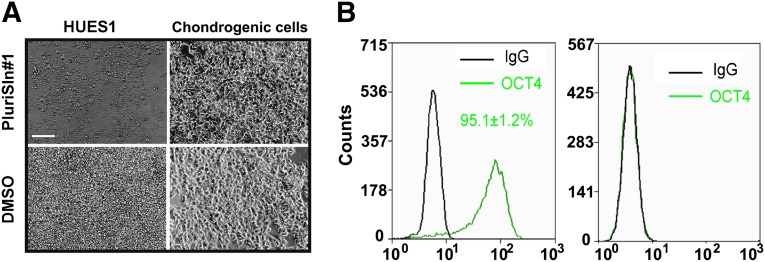
The stearoyl-CoA desaturase 1 (SCD1) inhibitor PluriSIn1 is reported to be selectively toxic to pluripotent cells at 20 μM [24]. We confirmed this toxicity with HUES1 cells, but at 20 μM it was also toxic to stage 3 cells. However, in further experiments with PluriSIn1 at 2.5 μM for 72 hours, it selectively killed HUES1 cells but was not toxic to the differentiated cells from stage 3, suggesting that pluripotency was lost during differentiation of the chondrogenic cells (Fig. 2A). Together with the lack of OCT4 positive cells detected using flow cytometry on day 14 of chondrogenesis, compared with 95% positive cells in HUES1 pluripotent cultures (Fig. 2B), this provided good evidence that the protocol was effective in generating differentiated cells with negligible residual pluripotent cells.
Figure 2.
Detection of residual pluripotent cells in human embryonic stem cell (hESC) chondrogenic cells. HUES1 and chondrogenic cells derived from these hESCs were treated with 2.5 μM PluriSIn1 or DMSO for 72 hours. (A): OCT4 expression in both types of cells was investigated with flow cytometry. (B): No significant difference was found for cells labeled with IgG control and OCT4. The results are the means ± SD (n = 3 experiments). Scale bar = 100 μm. Abbreviations: DMSO, dimethyl sulfoxide; NS, not significant.
iPSC Differentiation to Chondrocytes
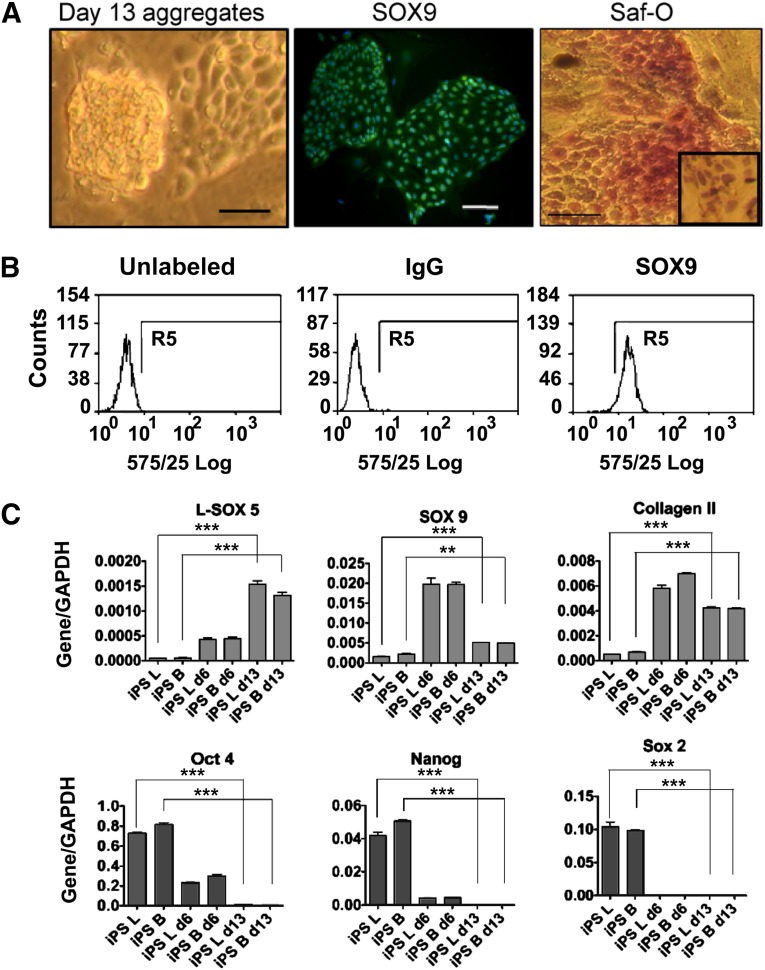
The revised differentiation protocol was also tested with two iPSC lines, ZK2012L and ZK2012B, generated in our group. The iPSCs were first evaluated for expression of pluripotency-associated markers and for their ability to generate EBs and teratomas, in each case giving cells of all three germ lineages (supplemental online Fig. 2C). These iPSCs in feeder-free culture maintained a hESC-like morphology with nuclear expression of the pluripotency associated markers OCT4, SOX2, and Nanog. By day 6 of directed differentiation, the cells had lost their hESC-like morphology and showed a flattened appearance, similar to hESCs at the same stage, and by day 13, they were more rounded with the morphology seen previously for chondrogenic precursors (Fig. 3A).
Figure 3.
Chondrogenic differentiation of iPSCs. iPSCs were differentiated into chondroprogenitors using the directed differentiation protocol. (A): At the end of the protocol, chondrogenic cell aggregation was observed, SOX9 expression was detected by immunofluorescence and the deposition of sulfated glycosaminoglycan was shown by Safranin O staining (inset shows cultures pretreated with chondroitinase ABC). Scale bars = 200 μm. (B): Flow cytometry study for SOX9 expression in chondrogenic cells from day 13 of the protocol. (C): Genes associated with pluripotency and chondrogenesis were measured by quantitative reverse transcription polymerase chain reaction. B and L indicate the two iPSC lines used for the experiment. The results are the means ± SD (n = 3). The levels of significance are depicted as follows: ∗∗, p < .01; ∗∗∗, p < .001. Abbreviations: d6, day 6; d13, day 13; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; iPS, induced pluripotent stem cells; Saf-O, Safranin O.
Chondrogenic differentiation of human iPSCs (hiPSCs) (between passages 12 and 25) was validated by immunostaining for SOX9, which showed nuclear localization (Fig. 3A), and by flow cytometry, the proportion of SOX9 positive cells at the end of the protocol was 96% ± 4.8% (n = 3) (Fig. 3B). At day 13, three-dimensional Safranin O-positive/chondroitinase-sensitive aggregates formed (Fig. 3A). Gene expression analyses of the iPSC lines showed chondrocyte-associated gene expression. COL2A1 increased eightfold between pluripotent hiPSCs and stage 3 (Fig. 3C), and there was a 12.7-fold upregulation in SOX9 between pluripotent iPSCs and cells at day 6, which fell to a 3.5-fold increase at day 13 (Fig. 3C). The expression of L-SOX5 showed a 30-fold increase between pluripotent iPSCs and cells at day 13 (Fig. 3C).
As with hESCs, in the iPSC lines the high expression of the pluripotency-associated genes OCT4, SOX2, and Nanog was lost during differentiation, being undetectable at day 13. This indicates loss of the pluripotent stem cell state (Fig. 3C). Collectively, the data showed evidence for the differentiation of iPSCs along the chondrogenic lineage.
Cartilage Regeneration in a Focal Osteochondral Defect
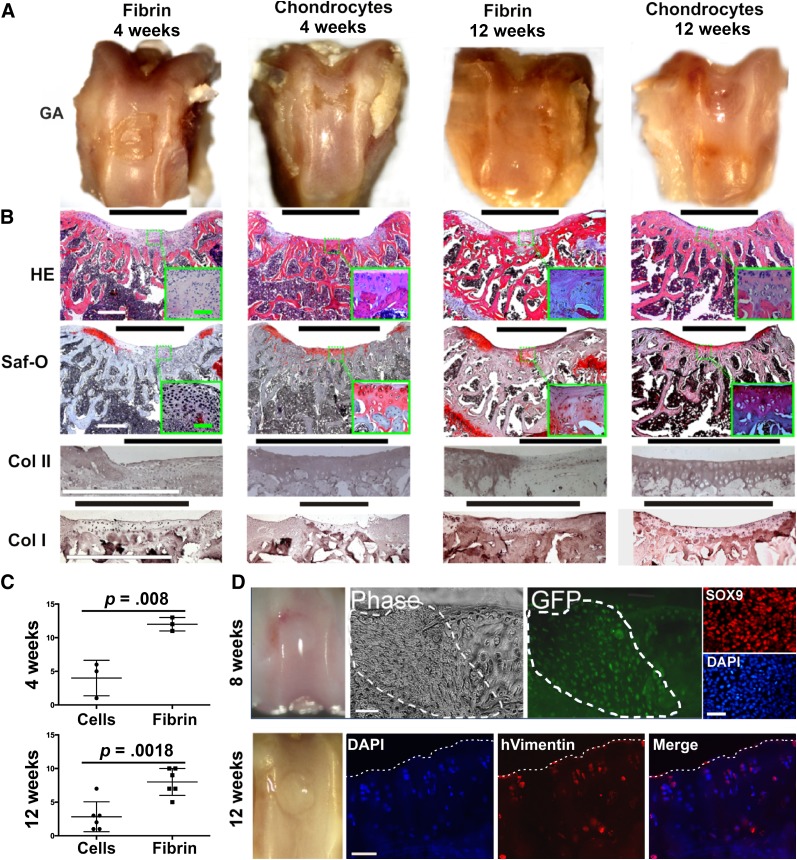
We assessed the regenerative potency of the chondroprogenitors derived from hESCs using an osteochondral defect model in the patellar groove of athymic RNU rats. To track the fate of the implanted cells, we labeled hESCs using lentivirus carrying an EF1 promoter-driven EGFP as reporter. When the cells were differentiated to chondroprogenitors and then encapsulated in fibrin gel, almost all cells were SOX9 positive after 3 days of culture. By 4 weeks, the defects treated with fibrin chondroprogenitors were partially repaired, mainly with flat, shiny, translucent tissue observed in the unfixed joint, but with some areas of irregular surface. This contrasted with fibrin-only controls, which contained minimal repair with rough and irregular tissue (Fig. 4A). At 8 weeks, the repair was even more complete in defects treated with fibrin chondroprogenitors, and at 12 weeks the fibrin chondroprogenitor-treated defects showed a smooth surface, which appeared similar to native cartilage. Fibrin-treated controls remained poorly repaired (Fig. 4A, 4D).
Figure 4.
In vivo cartilage formation promoted by chondrogenic cells derived from human embryonic stem cells. (A): RNU rats were killed at designated time points postimplantation, and the patella groove was assessed by macroscopic examination of the gross appearance after fixation. (B): Histological assessment of sections through knee cartilage using hematoxylin and eosin and Saf-O staining and by immunohistochemical staining for collagen I and II. (C): Cartilage repair scoring using Pineda’s system, which has a score, range from 0 (best) to 14 (worst). The results are the means ± SD (n = 3 animals for the 4 weeks group and n = 6 for the 12 weeks group; 18 animals in total) (D): GFP-positive cells (which expressed SOX9 before implantation, top right column) were detected 8 weeks after implantation of GFP-labeled chondrogenic cells (defect area outlined by dotted line). Human cells were detected 12 weeks after implantation by immunohistochemistry using anti-human vimentin antibody (surface of defect area shown by dotted line). Black bars = defect area; white bars = 500 μm; green bars = 100 μm. Abbreviations: Col, collagen; DAPI, 4′,6-diamidino-2-phenylindole; GA, gross appearance after fixation; GFP, green fluorescent protein; HE, hematoxylin and eosin; hVimentin, human vimentin; Saf-O, Safranin O.
Histology at 4 weeks after implantation of fibrin chondroprogenitors showed repair tissue with lacunae-containing cells resembling chondrocytes in a matrix with staining for sGAG and collagen II. In contrast, in controls treated with fibrin only, there was a high density of small fibroblastic cells lacking sGAG and collagen II staining. By 12 weeks, the sGAG and collagen II staining had increased in tissue repaired with fibrin chondroprogenitors, but the controls remained poorly stained. In contrast, in the repaired cartilage, collagen I staining was faint, similar to that in native cartilage, compared with the strong staining in the subchondral bone. This suggests the formation of hyaline cartilage from implanted chondrocytes derived from hESCs (Fig. 4B). We evaluated the repair using a histomorphological scoring system for cartilage [11, 12] and found significantly improved repair with hESC chondrogenic cells compared with fibrin-only controls after both 4 and 12 weeks of implantation (Fig. 4C). No tumor formation was found during the histological analysis.
To determine whether the human cells remained in the construct and were therefore potentially responsible for deposition of the cartilage matrix, cryosections of joint samples in which EGFP-labeled chondrogenic cells had been implanted were examined. At 8 weeks, EGFP-positive cells were present in the defect area including at the level of subchondral bone. The fate of implanted cells was also demonstrated with a human-specific anti-vimentin antibody, which stained many cells in the defect area at 12 weeks, confirming survival of viable human cells in the rat joint (Fig. 4D).
Discussion
In this study, we validated the previous reported protocol with further hESC and iPSC lines and refined the method to enhance the expression of SOX9 and collagen II, as well as to promote chondrogenic aggregation at the end of the protocol. We demonstrated that the hESC chondroprogenitors implanted in fibrin gel produced statistically significant enhanced repair of cartilage in vivo compared with fibrin scaffold alone in an osteochondral defect model in RNU rats. The repair tissue in the defect showed enhanced cartilage formation in the area where the hESC-derived chondrocytes were implanted, which stained strongly for sGAG and collagen II with negligible collagen I. This contrasted with the fibrin-only control, which showed negligible cartilage formation. In our scoring system, uninjured cartilage would score 0; thus the hESC-derived cartilage repair is not equivalent to uninjured cartilage but is significantly better the repair of defects treated with fibrin alone. Importantly, we also showed that pluripotent cells were undetectable in the hESC chondrogenic cell population, and no evidence of abnormal tissue formation or tumors was detected after 12 weeks in vivo. These results thus mark an important step forward in supporting further development toward clinical translation. Clinical grade hESCs and their defined differentiation into chondrocytes offer the twin advantages of generating cells of reproducible repair potential and the possibility of banking large numbers of cells for delivery to many patients, because hESCs retain the capacity for almost unlimited proliferation (self-renewal). Although conclusive evidence is not yet in place, there are suggestions that cartilage repair may be feasible using allogeneic cells with limited tissue matching [25], opening the way to more widespread use of hESCs as a therapeutic cell source. In our protocol, hESCs differentiate rapidly and efficiently within 14 days with a greater than eightfold cell expansion [9]. Supported by our in vivo results, this shows that the protocol has strong potential for development to generate large numbers of chondrogenic cells appropriate for clinical use. The development of robust, chemically defined differentiation protocols combined with clinical grade hESC lines makes future application feasible [26].
In this study, we have also shown that iPSCs also responded to our protocol. This is important because these cells may also be used to make patient-specific chondrocytes for treatment in the future. However, the low efficiency of reprogramming to pluripotency, concerns regarding their differences from authentic hESCs [27], and issues of retention of differentiated epigenetic memory are additional hurdles that need to be overcome. It was notable that the iPSCs showed a slightly reduced response to hESCs in the later chondrogenic phase of our protocol. This suggests that conditions may not be optimal for iPSCs, or this could reflect the process of reprogramming including epigenetic memory. It is also possible that at low passage number, the cells may be less responsive to differentiation signals, although we have not found this with hESCs.
Because our protocol mimics intrinsic human chondrocyte differentiation, it will also be invaluable in studying the processes involved in cartilage development in vitro and developing human disease models for conenital skeletal defects. The application of the protocol to iPSCs, pluripotent cells of quite different origin from hESCs, illustrates the intrinsic nature of the signals involved. Up to now, there has been limited research on the derivation of chondrocytes from iPSCs. Differentiation of iPSCs derived from osteoarthritic chondrocytes was reported using retroviral transduction of transforming growth factor β1 (TGFβ1), combined with coculture with existing chondrocytes on an alginate matrix [28]. Although this combined method improved on coculture or TGFβ1 transduction alone, the use of primary chondrocytes makes the process variable and undefined. The direct transdifferentiation of mouse adult dermal fibroblasts into chondrocytes has also been attempted through the retroviral expression of two reprogramming factors (c-Myc and Klf4) and one chondrogenic factor, SOX9 [29]. This eliminated the need for iPSCs; however, the resultant chondrogenic cultures showed karyotypic instability, tumor formation, and hypertrophic cartilage formation. Our differentiation protocol represents a major improvement in the efficient derivation of chondrogenic cells under defined conditions from pluripotent cells.
Chondrocyte adhesion to substrates in monolayer culture was reported to induce actin stress fiber formation and loss of SOX9 expression, so reduced substrate adhesion in cell aggregates may influence the number of cells expressing SOX9 and its level of expression [30]. The evidence of a type II collagen and chondroitin sulfate-rich matrix at the end of the protocol clearly shows that the expression of SOX9 in the hESC-derived chondroprogenitors is sufficient to drive cartilage matrix protein production.
Conclusion
We have shown that hESC chondrogenic cells generated using a chemically defined serum-free protocol can generate cartilage repair in vivo, proof of principle for using hESCs as a cell source for cartilage tissue engineering. The protocol provides a system that will enable detailed analysis of the mechanisms driving chondrogenesis in health and disease and a platform for diverse research applications.
Supplementary Material
Acknowledgments
A.C. and S.J.K. thank Arthritis Research UK (Grant R110927) and BIS UK, and Z.K. and R.J.L. thank the Biomedical Research Council for providing financial support for this study. Z.K. and S.J.K. thank the Wellcome Trust Institutional Strategic Support Fund (Award 097820). J.P. and S.L. thank National Natural Science Foundation of China (Grants 31240048 and 30930092) for financial support. We are grateful to Nicola Bates for technical support, Sara Dunn for osteoarthritic cartilage, and Dr. Jinpei Ye for the derivation of the MAN7 hESC line.
Author Contributions
A.C.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; Z.K.: conception and design, collection and assembly of data, data analysis and interpretation, final approval of manuscript; J.P.: collection and assembly of data, data analysis and interpretation, final approval of manuscript; S.L.: conception and design, data analysis and interpretation, final approval of manuscript; R.J.L.: conception and design, final approval of manuscript; T.E.H.: conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript; S.J.K.: conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Brittberg M, Lindahl A, Nilsson A, et al. Treatment of deep cartilage defects in the knee with autologous chondrocyte transplantation. N Engl J Med. 1994;331:889–895. doi: 10.1056/NEJM199410063311401. [DOI] [PubMed] [Google Scholar]
- 2.Filardo G, Kon E, Berruto M, et al. Arthroscopic second generation autologous chondrocytes implantation associated with bone grafting for the treatment of knee osteochondritis dissecans: Results at 6 years. Knee. 2012;19:658–663. doi: 10.1016/j.knee.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Kang SW, Yoo SP, Kim BS. Effect of chondrocyte passage number on histological aspects of tissue-engineered cartilage. Biomed Mater Eng. 2007;17:269–276. [PubMed] [Google Scholar]
- 4.Hettrich CM, Crawford D, Rodeo SA. Cartilage repair: Third-generation cell-based technologies: Basic science, surgical techniques, clinical outcomes. Sports Med Arthrosc. 2008;16:230–235. doi: 10.1097/JSA.0b013e31818cdc98. [DOI] [PubMed] [Google Scholar]
- 5.Pelttari K, Winter A, Steck E, et al. Premature induction of hypertrophy during in vitro chondrogenesis of human mesenchymal stem cells correlates with calcification and vascular invasion after ectopic transplantation in SCID mice. Arthritis Rheum. 2006;54:3254–3266. doi: 10.1002/art.22136. [DOI] [PubMed] [Google Scholar]
- 6.Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 7.Wakitani S, Okabe T, Horibe S, et al. Safety of autologous bone marrow-derived mesenchymal stem cell transplantation for cartilage repair in 41 patients with 45 joints followed for up to 11 years and 5 months. J Tissue Eng Regen Med. 2011;5:146–150. doi: 10.1002/term.299. [DOI] [PubMed] [Google Scholar]
- 8.Stolzing A, Jones E, McGonagle D, et al. Age-related changes in human bone marrow-derived mesenchymal stem cells: Consequences for cell therapies. Mech Ageing Dev. 2008;129:163–173. doi: 10.1016/j.mad.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Oldershaw RA, Baxter MA, Lowe ET, et al. Directed differentiation of human embryonic stem cells toward chondrocytes. Nat Biotechnol. 2010;28:1187–1194. doi: 10.1038/nbt.1683. [DOI] [PubMed] [Google Scholar]
- 10.Baxter MA, Camarasa MV, Bates N, et al. Analysis of the distinct functions of growth factors and tissue culture substrates necessary for the long-term self-renewal of human embryonic stem cell lines. Stem Cell Res (Amst) 2009;3:28–38. doi: 10.1016/j.scr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Pineda S, Pollack A, Stevenson S, et al. A semiquantitative scale for histologic grading of articular cartilage repair. Acta Anat (Basel) 1992;143:335–340. doi: 10.1159/000147272. [DOI] [PubMed] [Google Scholar]
- 12.Orth P, Zurakowski D, Wincheringer D, et al. Reliability, reproducibility, and validation of five major histological scoring systems for experimental articular cartilage repair in the rabbit model. Tissue Eng Part C Methods. 2012;18:329–339. doi: 10.1089/ten.TEC.2011.0462. [DOI] [PubMed] [Google Scholar]
- 13.Ng LJ, Wheatley S, Muscat GE, et al. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- 14.Bell DM, Leung KK, Wheatley SC, et al. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 15.Lefebvre V, Huang W, Harley VR, et al. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstin E, Hooper ML, Grandcha S, et al. Alkaline-phosphatase activity in mouse teratoma. Proc Natl Acad Sci USA. 1973;70:3899–3903. doi: 10.1073/pnas.70.12.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu TM, Guo XM, Tan HS, et al. Zinc-finger protein 145, acting as an upstream regulator of SOX9, improves the differentiation potential of human mesenchymal stem cells for cartilage regeneration and repair. Arthritis Rheum. 2011;63:2711–2720. doi: 10.1002/art.30430. [DOI] [PubMed] [Google Scholar]
- 18.Han Y, Lefebvre V. L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer. Mol Cell Biol. 2008;28:4999–5013. doi: 10.1128/MCB.00695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefebvre V, Behringer RR, de Crombrugghe B. L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage. 2001;9(suppl A):S69–S75. doi: 10.1053/joca.2001.0447. [DOI] [PubMed] [Google Scholar]
- 20.Furumatsu T, Tsuda M, Yoshida K, et al. Sox9 and p300 cooperatively regulate chromatin-mediated transcription. J Biol Chem. 2005;280:35203–35208. doi: 10.1074/jbc.M502409200. [DOI] [PubMed] [Google Scholar]
- 21.Dvir-Ginzberg M, Gagarina V, Lee EJ, et al. Regulation of cartilage-specific gene expression in human chondrocytes by SirT1 and nicotinamide phosphoribosyltransferase. J Biol Chem. 2008;283:36300–36310. doi: 10.1074/jbc.M803196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leijten JCH, Emons J, Sticht C, et al. Gremlin 1, frizzled-related protein, and Dkk-1 are key regulators of human articular cartilage homeostasis. Arthritis Rheum. 2012;64:3302–3312. doi: 10.1002/art.34535. [DOI] [PubMed] [Google Scholar]
- 23.Kielty CM, Kwan AP, Holmes DF, et al. Type X collagen, a product of hypertrophic chondrocytes. Biochem J. 1985;227:545–554. doi: 10.1042/bj2270545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-David U, Gan QF, Golan-Lev T, et al. Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell. 2013;12:167–179. doi: 10.1016/j.stem.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 25.Sato M, Uchida K, Nakajima H, et al. Direct transplantation of mesenchymal stem cells into the knee joints of Hartley strain guinea pigs with spontaneous osteoarthritis. Arthritis Res Ther. 2012;14:R31. doi: 10.1186/ar3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacquet L, Stephenson E, Collins R, et al. Strategy for the creation of clinical grade hESC line banks that HLA-match a target population. EMBO Mol Med. 2013;5:10–17. doi: 10.1002/emmm.201201973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilic J, Izpisua Belmonte JC. Concise review: Induced pluripotent stem cells versus embryonic stem cells: Close enough or yet too far apart? Stem Cells. 2012;30:33–41. doi: 10.1002/stem.700. [DOI] [PubMed] [Google Scholar]
- 28.Wei Y, Zeng W, Wan R, et al. Chondrogenic differentiation of induced pluripotent stem cells from osteoarthritic chondrocytes in alginate matrix. Eur Cell Mater. 2012;23:1–12. doi: 10.22203/ecm.v023a01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiramatsu K, Sasagawa S, Outani H, et al. Generation of hyaline cartilaginous tissue from mouse adult dermal fibroblast culture by defined factors. J Clin Invest. 2011;121:640–657. doi: 10.1172/JCI44605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tew SR, Hardingham TE. Regulation of SOX9 mRNA in human articular chondrocytes involving p38 MAPK activation and mRNA stabilization. J Biol Chem. 2006;281:39471–39479. doi: 10.1074/jbc.M604322200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.