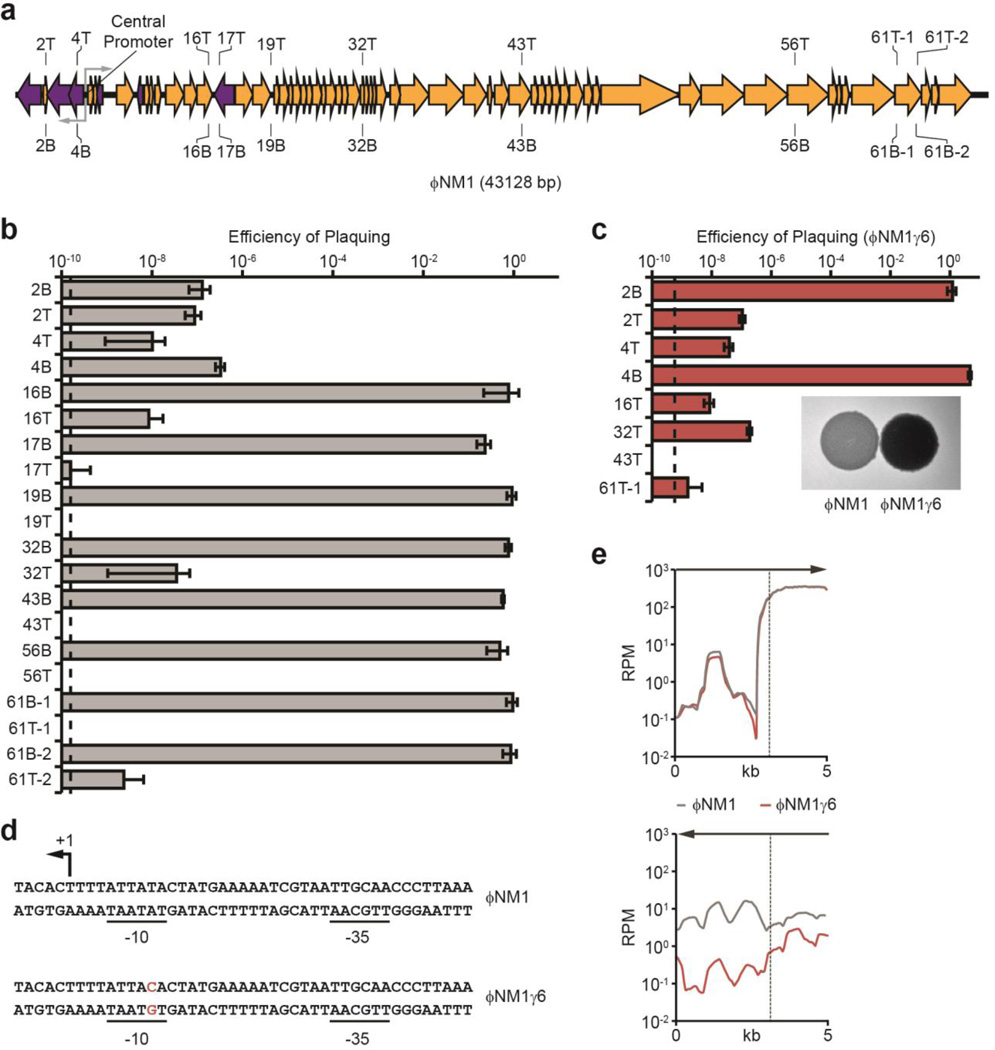
Fig. 2. Transcription of target sequences is required for type III-A CRISPR immunity.
a, Schematic diagram of the ΦNM1 genome and the position of targets used in this study. T, crRNA anneals to the top strand; B, bottom strand. Gray arrows represent the ΦNM1 central promoter driving divergent transcription. b, Immunity against ΦNM1 infection provided by spacers targeting the phage regions shown in a. Dotted line indicates the limit of detection for the assay. c, Immunity against ΦNM1γ6 infection. Inset; comparison of lysis phenotypes for ΦNM1 (turbid) and ΦNM1γ6 (clear), representative of four technical replicates. d, Leftward promoter consensus sequences at the ΦNM1 and ΦNM1γ6 central promoter. The ΦNM1γ6 mutation in the −10 element is shown in red. The putative transcription start site is noted (+1). e, Comparison of phage transcription profiles from cells infected with ΦNM1 (gray line) or ΦNM1γ6 (red line), 15 minutes post-infection. Phage-derived transcripts are plotted in reads per million total-mapped reads (RPM) relative to their position on the genome; arrows indicate the direction of transcription plotted in each graph; the vertical dotted line marks the position of the central promoter. Error bars: mean ± s.d. (n=3).