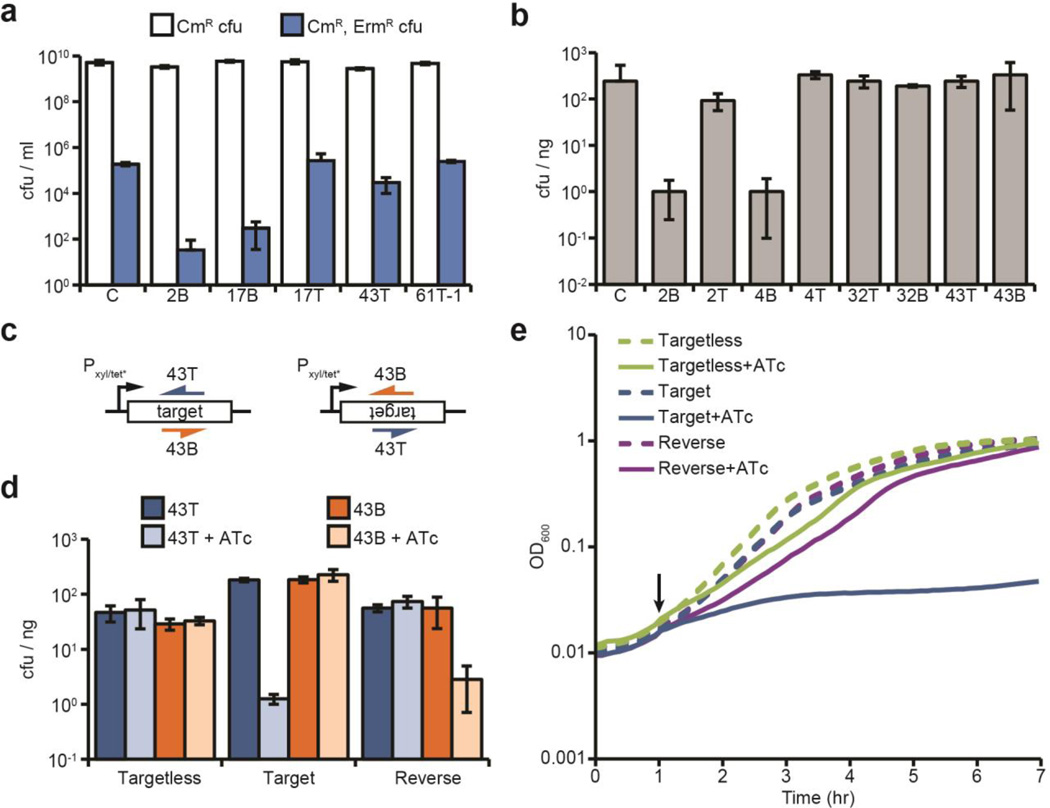
Fig. 3. Conditional tolerance is achieved via transcription-dependent CRISPR-Cas targeting.
a, ΦNM1-ErmR lysogenization for additional spacers. C, pGG3 non-targeting control. b, Transformation of ΦNM1-lysogenic competent cells with CRISPR-Cas plasmids containing different spacers (transformation efficiency is measured as cfu/ng of plasmid DNA). C, pGG3 non-targeting control. c, Integration of the 43T/B ΦNM1 target region into the chromosome of S. aureus. Target sequences (inserted in both forward and reverse orientations) are under the control of the tetracycline-inducible promoter Pxyl/tet*. The 43T/B crRNAs are shown annealing to either the top or bottom strands. d, Transformation of both strains shown in c, as well as an isogenic control strain lacking the target insertion, with CRISPR-Cas plasmids containing spacers 43T or 43B. Transformants were plated on selective plates with or without anhydrotetracycline (ATc) for induction of the Pxyl/tet* promoter. e, Growth curve of strains shown in d expressing the spacer 43T CRISPR-Cas system, in the presence or absence of ATc addition at the indicated timepoint (black arrow). Error bars: mean ± s.d. (n=3).