Extended Data Figure 1. Characterization of spacer 32T isolates lysogenized with ΦNM1-ErmR.
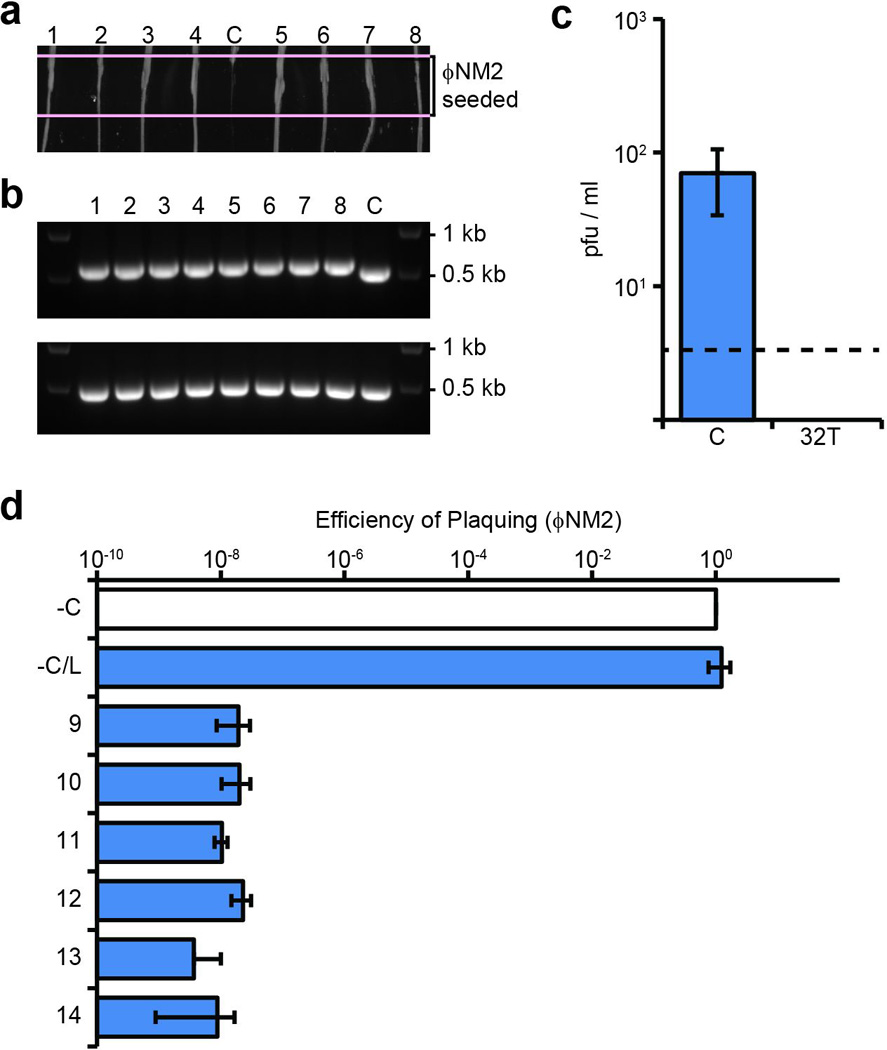
a, ΦNM2 sensitivity assay. Eight randomly selected ΦNM1-ErmR lysogen clones were re-streaked through the indicated ΦNM2-seeded region from top to bottom (1–8); C, sensitive ΦNM1-ErmR lysogen harboring the pGG3 control plasmid. b, PCR amplification of the CRISPR array (upper panel) and spacer 32T target region (lower panel) for the strains tested in a. The pGG3 control lysogen (C) lacks a phage-targeting spacer in its CRISPR array. 1 kb and 0.5 kb size markers are indicated. All 8 PCR products for the target region were sequenced by the Sanger method and no mutations were found (data not shown). c, Plaque-forming potential of filtered supernatants from spacer 32T lysogen overnight cultures inoculated in triplicate. Plaque-forming units (pfu) were enumerated on soft agar lawns of RN4220 harboring either the pGG3 control (C) or spacer 32T CRISPR plasmids. Dotted line represents the limit of detection for this assay. d, ΦNM2 plaquing efficiency on soft agar lawns of an additional six randomly selected ΦNM1-ErmR lysogen clones isolated during infection of RN4220/spacer 32T (9–14); a ΦNM1-ErmR lysogen harboring the pGG3 control plasmid was also tested (−C/L). Plaquing efficiency on the non-lysogenic indicator strain harboring pGG3 is shown for comparison (−C). Error bars: mean ± s.d. (n=3). Panels a and b represent single experiments performed for 8 of 32 isolates.
