Extended Data Figure 4. Type II CRISPR-Cas targeting in S. aureus prevents both lytic and lysogenic infection.
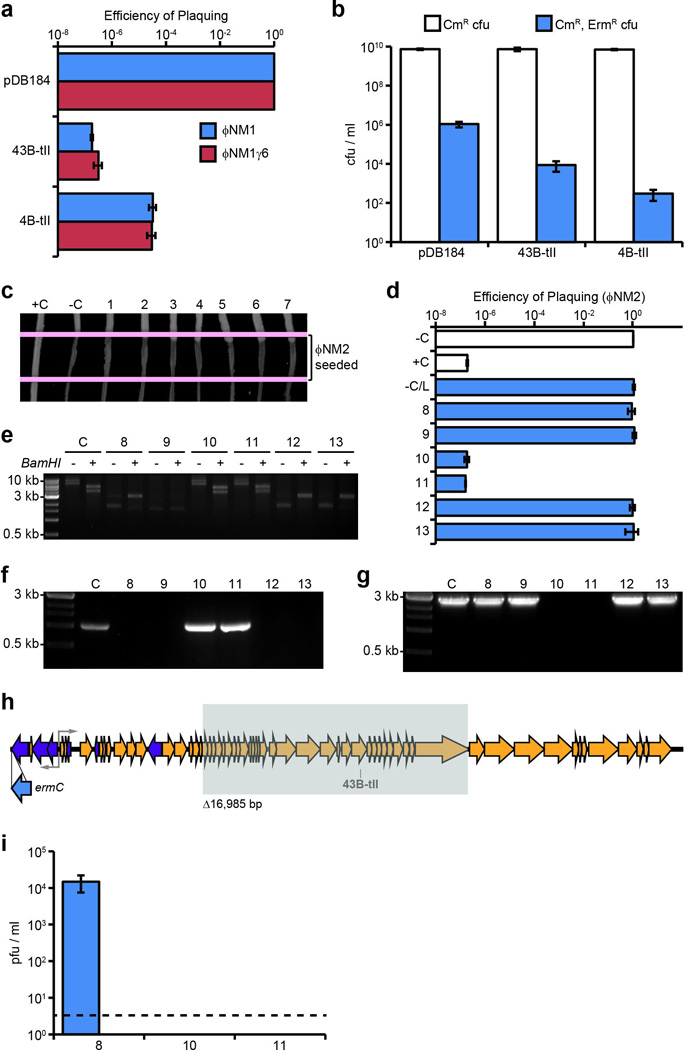
a, Plaquing efficiency of ΦNM1 and ΦNM1γ6 on lawns of RN4220 harboring type II-A CRISPR-Cas plasmids as indicated. The parental vector, pDB184, serves as a non-targeting control. b, ΦNM1-ErmR lysogenization of RN4220 harboring either the spacer 43B-tII, 4B-tII, or non-targeting type II-A CRISPR plasmids. c, ΦNM2 sensitivity assay for seven randomly selected ΦNM1-ErmR lysogen clones isolated during infection of RN4220/spacer 43B-tII (1–7). For comparison, a resistant non-lysogen harboring the spacer 43B-tII plasmid and a sensitive lysogen harboring the pDB184 plasmid were included as controls (respectively, C+ and C−). Picture represents a single experiment for 7 of 22 isolates. d, ΦNM2 plaquing efficiency on soft agar lawns for an additional six randomly selected ΦNM1-ErmR lysogen clones isolated during infection of RN4220/spacer 43B-tII (8–13); a ΦNM1-ErmR lysogen harboring the pDB184 plasmid is also tested (−C/L). For comparison, plaquing efficiency of ΦNM2 on the non-lysogenic indicator strain harboring pDB184 or the targeting spacer 43B-tII plasmid are also shown (−C and +C, respectively). e, Agarose gel electrophoresis of plasmid DNA purified from isolates 8–13 and the parental spacer 43B-tII strain (C). +/− indicate the presence or absence of treatment with the BamHI restriction enzyme which produces 2 bands for the wild type spacer 43B-tII plasmid: 5367 bp and 3972 bp. Size markers correspond to 10 kb, 3 kb, and 0.5 kb bands of the 1kb DNA ladder from NEB. f, Colony PCR spanning the type II CRISPR array for isolates 8–13. Spacer 43B-tII plasmid DNA was used as a template for the control (C). 3 kb and 0.5 kb size markers are indicated. g, Colony PCR spanning the target region for isolates 8–13 and a ΦNM1-ErmR lysogen harboring the pDB184 control plasmid (C). Isolates # 10 and 11 harbor identical deletions within the prophage that remove the target region (see below). 3 kb and 0.5 kb size markers are indicated. The presence of attL and attR prophage integration arms was also verified independently for each isolate using PCR (data not shown). h, Location of the 16,985 bp deletion identified within the prophage harbored by isolates # 10 and 11 (shaded gray box). The location and orientation of the ermC insertion cassette is also shown (blue arrow). Deletion was mapped by primer walking. An ~9.1 kb product spanning the deletion was ultimately amplified using primers oGG6 and oGG241, and the deletion junction was sequenced by the Sanger method using oGG245. A perfect 14 bp direct repeat micro-homology flanks the deletion. i, Plaque-forming potential of overnight culture supernatants from isolates # 8, 10, and 11. Supernatants were plated by the soft agar method with RN4220 cells harboring the non-targeting pDB184 control plasmid as an indicator strain. Supernatants were also plated with spacer 43B-tII targeting lawns, yielding no detectable pfu. Isolate 8 appears to exhibit wild type levels of spontaneous prophage induction (compare to pGG3 control in Fig. 4a). No plaque-forming units were detected from the supernatants of isolates # 10 and 11 whatsoever, presumably resulting from their deletion of genes essential for prophage induction, including the ORF 43 major capsid protein. Dotted line represents the limit of detection for this assay. Error bars: mean ± s.d. (n=3). Panels e through g represent single experiments for 6 of 22 isolates.
