Figure 6.
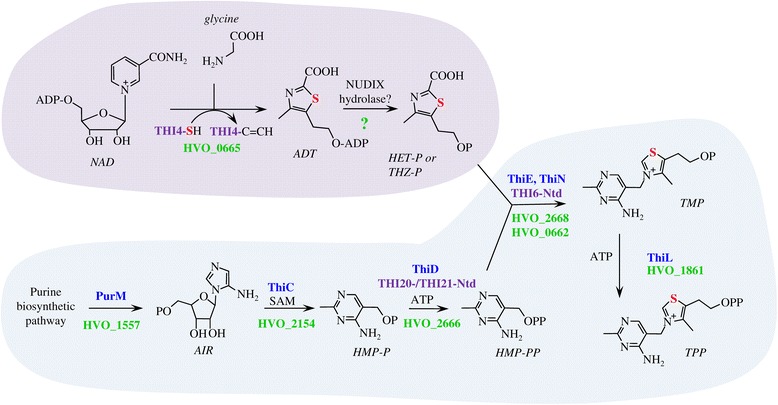
The pathway for de novo biosynthesis of thiamine in the archaeon Haloferax volcanii is an apparent hybrid of bacterial and yeast pathways. Yeast and bacterial like steps are shaded by purple and blue, respectively, with associated enzymes of thiamine biosynthetic indicated in text with similar color coding. Hfx. volcanii homologs are indicated by gene locus tag in green. Question mark (?) designated enzyme is yet unassigned. THI4-SH specifies the catalytic cysteine side chain. THI4-C = CH indicates the dehydroalanine form of the enzyme after sulfur transfer. The sulfur atom associated with formation of the thiazole ring is highlighted in red. In the Hfx. volcanii model for thiamine biosynthesis, the thiazole moiety of thiamine (4-methyl-5-(β-hydroxyethyl)thiazole phosphate; HET-P or THZ-P) is generated by a yeast-like mechanism. HvThi4 (HVO_0665) converts nicotinamide adenine dinucleotide (NAD) and glycine to ADP-thiazole (ADT), which predicted to be hydrolyzed to THZ-P by a yet to be identified NUDIX-type hydrolase [29]. The remaining steps of thiamine biosynthesis are related to bacterial systems. In particular, the PurM-like AIR synthetase HVO_1557 is predicted to form 5-amino-1-(5-phospho-D-ribosyl)imidazole (synonym 5-aminoimidazole ribotide; AIR), which serves as a substrate for a ThiC-like S-adenosyl-methionine (SAM)-dependent HMP-P synthase (HVO_2154) in the generation of 4-amino-hydroxymethyl-2-methylpyrimidine phosphate (HMP-P). Once generated, HMP-P is phosphorylated by a bacterial ThiD-type HMP-P kinase (HVO_2666) which is also conserved in the N-terminal domains (Ntd) of yeast THI20 and THI21. Thiamine-phosphate synthase homologs of the ThiE-type (HVO_2668) and ThiN-type (HVO_0662) are predicted to condense THZ-P with HMP-PP to form thiamine monophosphate (TMP). TMP is then phosphorylated to TPP by a proposed bacterial ThiL-type thiamine-monophosphate kinase (HVO_1861).
