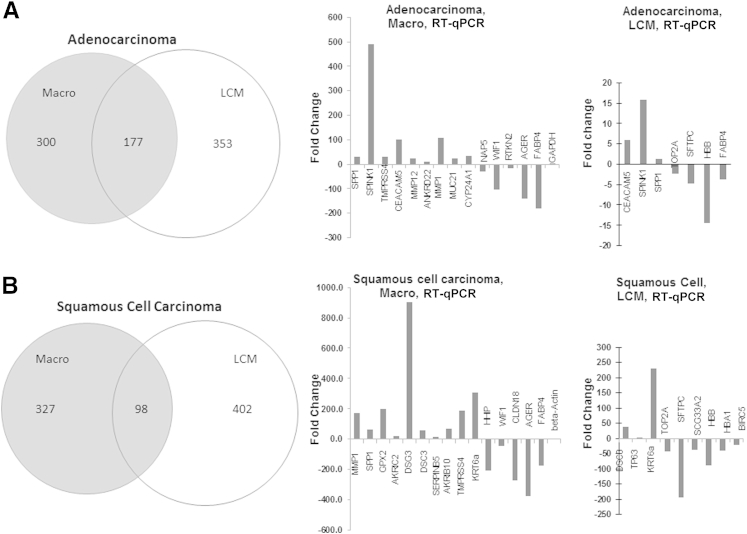
Figure 3.
Validation of adenocarcinoma-specific (A) and squamous cell carcinoma–specific (B) DE transcripts in Macro versus LCM specimens. A: The analysis revealed 177 T versus NT DE transcripts in common between the Macro and LCM platforms (approximately 27%), shown in the Venn diagram. Validation of T versus NT microarray-based differential expression by quantitative real-time RT-PCR (RT-qPCR) in Macro and LCM specimens. PCR-based assays of top microarray hits were evaluated using RNA-specific RT-qPCR on the same adenocarcinoma sample, as described under Materials and Methods, are shown in the graphs. The values for Macro tissue sets were concordant (ie, in the same direction, up-regulated or down-regulated) with LCM microarray values (Table 2) for the selected top microarray hits. Such concordance was also found with RT-qPCR validation, although of course the actual lists of the most dysregulated genes differed between Macro and LCM specimens. B: The analysis revealed 98 T–NT DE transcripts in common (approximately 23%) between the Macro and LCM platforms (shown in Venn diagram). Validation of squamous cell carcinoma T versus NT microarray-based differential expression by RT-qPCR in Macro and LCM specimens is shown in the graphs. Assays of top microarray hits were evaluated using an RNA-specific RT-qPCR on the same sample, as described under Materials and Methods. The values for Macro tissue sets were concordant with LCM microarray values (Table 1, Table 2, Table 3) for the selected top microarray hits. Such concordance was largely true of the LCM RT-qPCR validation for LCM microarray top hits, the two exceptions being TOP2A and BIRC5, which were up-regulated on the microarray but down-regulated in the RT-qPCR. Also, TP63 was up-regulated only 1.6-fold, albeit in the same qualitative direction as the microarray data. Again, the actual lists of the most dysregulated genes differed between Macro and LCM specimens. Data are expressed as mean FC (T versus NT) values, scaled to RNA-specific amplification of a housekeeping gene (GAPDH) (A) or to parallel RNA-specific amplification of a housekeeping transcript (β-actin) (B) not confounded by pseudogenes.