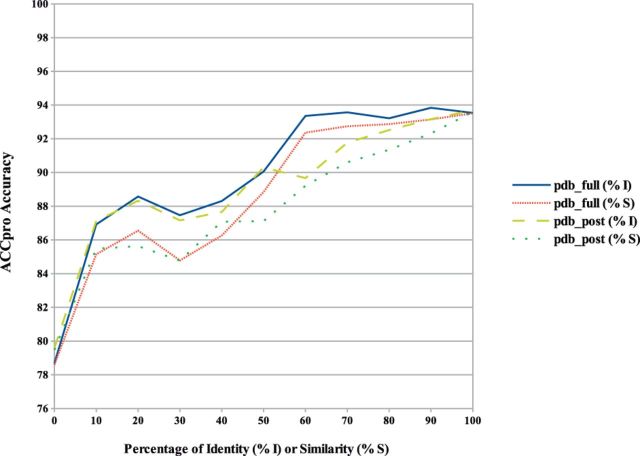
Fig. 7.
ACCpro prediction accuracy on the pdb_full and pdb_post datasets calculated as a function of the percentage of sequence identity or similarity (BLAST positive substitutions) with proteins found in the PDB. Cases where no similar sequence is found in the PDB for a given residue position, and thus predicted without using sequence-based structural similarity, are included in the 0% sequence identity or similarity case