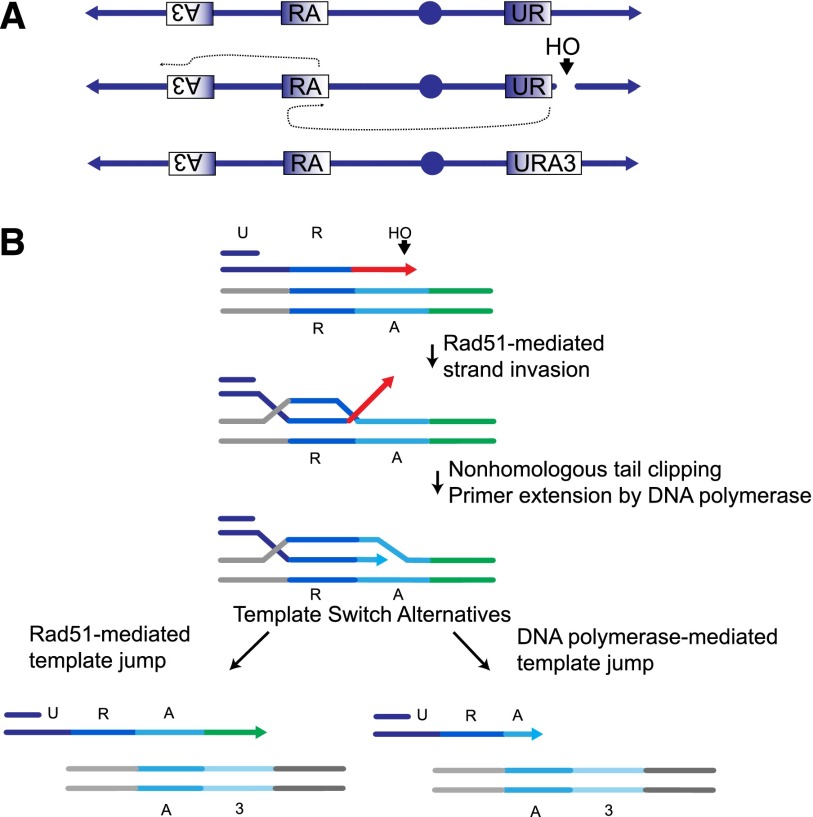
Figure 1.
Mechanisms of chromosome rearrangements via BIR and template switching. (A) Genetic assay to measure the frequencies of template switching following a DSB. The assay system is based on the 804-bp URA3 gene from S. cerevisiae, which was split into three overlapping segments, each sharing 300-bp homology, that we call “UR,” “RA,” and “A3” (see also Supplemental Fig. 1). A DSB is induced by the galactose-inducible HO endonuclease adjacent to the UR locus, located at the CAN1 locus in a nonessential terminal region of chromosome 5 (Chr 5); this break can be repaired by a BIR mechanism using, as the template, the RA sequence located in the middle of the opposite chromosome arm. Chr 5 is shown in reversed orientation from the conventional representation. Template switching from RA to A3, located 50 kb more distally on the same chromosome arm, will result in the creation of a functional URA3 gene as part of a nonreciprocal translocation that can be screened by selection on media lacking uracil. (B). Examining the mechanism of BIR and template switching. HO endonuclease creates a DSB next to the UR sequence. Following strand invasion into the RA template (dark-blue line followed by cyan line), clipping of the nonhomologous 3′ tail (red line) is a prerequisite for DNA synthesis. Template switching after the initial strand invasion can proceed by two alternative pathways. (Left) DNA synthesis proceeds past the RA template, and the subsequent template switch intermediate (cyan followed by green) carries a nonhomologous 3′ tail (green line). Subsequent DNA synthesis using the A3 sequence requires clipping of the nonhomologous 3′ tail (green line). (Right) Alternatively, a template switch into the A3 sequence (cyan followed by light-blue line) occurs during synthesis within the RA template. Subsequent DNA synthesis using the A3 sequence does not require tail clipping. With regards to tail clipping, the alternative shown on the left is similar to the first strand invasion event. In contrast, the alternative shown on the right is mechanistically different from the first strand invasion event, as it does not require tail clipping. The two alternatives are distinguished by examining the microhomology usage of the 3′ ends (see the text for details). Gray lines represent adjacent, nonhomologous, flanking sequences.