The effector complex of the Cmr/type III-B CRISPR–Cas system cleaves RNAs recognized by the crRNA of the complex and includes six protein subunits of unknown functions. Hale et al. used reconstituted Pyrococcus furiosus Cmr complexes and found that Cmr3 recognizes the signature crRNA tag sequence (and depends on protein–protein interactions with Cmr2, Cmr4, and Cmr5), each Cmr4 subunit mediates a target RNA cleavage, and Cmr1 and Cmr6 mediate an essential interaction between the 3′ region of the crRNA and the target RNA.
Keywords: Cmr, crRNA, CRISPR, Cas, RNAi, Pyrococcus furiosus
Abstract
The effector complex of the Cmr/type III-B CRISPR (clustered regularly interspaced short palindromic repeat)–Cas (CRISPR-associated) system cleaves RNAs recognized by the CRISPR RNA (crRNA) of the complex and includes six protein subunits of unknown functions. Using reconstituted Pyrococcus furiosus Cmr complexes, we found that each of the six Cmr proteins plays a critical role in either crRNA interaction or target RNA capture. Cmr2, Cmr3, Cmr4, and Cmr5 are all required for formation of a crRNA-containing complex detected by native gel electrophoresis, and the conserved 5′ repeat sequence tag and 5′-OH group of the crRNA are essential for the interaction. Interestingly, capture of the complementary target RNA additionally requires both Cmr1 and Cmr6. In detailed functional studies, we determined that P. furiosus Cmr complexes cleave target RNAs at 6-nucleotide (nt) intervals in the region of complementarity, beginning 5 nt downstream from the crRNA tag and continuing to within ∼14 nt of the 3′ end of the crRNA. Our findings indicate that Cmr3 recognizes the signature crRNA tag sequence (and depends on protein–protein interactions with Cmr2, Cmr4, and Cmr5), each Cmr4 subunit mediates a target RNA cleavage, and Cmr1 and Cmr6 mediate an essential interaction between the 3′ region of the crRNA and the target RNA.
CRISPR (clustered regularly interspaced short palindromic repeat)–Cas (CRISPR-associated) systems confer bacteria and archaea with the ability to acquire immunity to viruses and plasmids (Terns and Terns 2011; Westra et al. 2012; Sorek et al. 2013; Barrangou and Marraffini 2014; van der Oost et al. 2014). Upon infection, short fragments of invading DNA are captured and incorporated into discrete host genome loci termed CRISPRs, which consist of multiple copies of a short repeat sequence separated by the short invader-derived sequences (Bolotin et al. 2005; Mojica et al. 2005; Pourcel et al. 2005). CRISPR transcripts are processed into small CRISPR RNAs (crRNAs) that contain an invader-derived guide sequence and sequence from the flanking repeats (Brouns et al. 2008; Hale et al. 2008, 2009). crRNAs are loaded into effector complexes that can recognize and silence corresponding RNAs or DNAs. Modules of Cas proteins function in invader sequence acquisition and crRNA biogenesis as well as target destruction (Haft et al. 2005; Makarova et al. 2011a).
Bioinformatic analyses have identified three broad types of CRISPR–Cas systems, each with distinct types of crRNA-containing effector complexes (Makarova et al. 2011b). In the prototypic type I system found in Escherichia coli, multiple Cas proteins form a crRNA-containing complex that recognizes complementary DNA targets and recruits the Cas3 nuclease for target destruction (Brouns et al. 2008; Jore et al. 2011; Sinkunas et al. 2011, 2013; Wiedenheft et al. 2011a,b; Sashital et al. 2012; Westra et al. 2012; Mulepati and Bailey 2013; Hochstrasser et al. 2014; Huo et al. 2014; Jackson et al. 2014; Mulepati et al. 2014; Zhao et al. 2014). The prototypic type II effector complex also cleaves DNA targets but consists of a single protein (Cas9) associated with two essential RNAs (a crRNA guide and a tracrRNA cofactor) (Garneau et al. 2010; Deltcheva et al. 2011; Gasiunas et al. 2012; Jinek et al. 2012). Type III effector complexes (Hale et al. 2009, 2012; Spilman et al. 2013; Zhang et al. 2012; Staals et al. 2013) include multiple Cas proteins. The type III-B or Cmr complexes, characterized in Pyrococcus furiosus (Hale et al. 2009, 2012; Spilman et al. 2013), Sulfolobus solfataricus (Zhang et al. 2012), and Thermus thermophilus (Staals et al. 2013), cleave RNA targets. The type III-A or Csm subtype system targets DNA (Marraffini and Sontheimer 2008; Millen et al. 2012). The type I and type III effector complexes appear to be distantly related: The protein subunits of both complexes are members of common superfamilies that are found in strikingly similar arrangements within the complexes (Spilman et al. 2013).
The functions of the individual proteins in multisubunit CRISPR–Cas complexes are largely unknown. Type III-B Cmr complexes from P. furiosus consist of the six Cmr proteins, Cmr1–6, and a crRNA comprised of 8 nucleotides (nt) of CRISPR sequence at the 5′ end (the signature 5′ tag) and typically either 37 or 31 nt of invader-derived guide sequence (45- and 39-nt crRNAs) (Hale et al. 2008, 2009, 2012). (The Cmr complex from T. thermophilus has a similar composition [Staals et al. 2013].) Cmr1, Cmr4, and Cmr6 are members of the Cas7 superfamily of proteins found in both type I and type III systems (Makarova et al. 2011a). Cmr3 is a member of the Cas5 protein superfamily, also found in both type I and type III systems (Makarova et al. 2011a). All four of these Cmr proteins (Cmr1, Cmr3, Cmr4, and Cmr6) contain a distinct ferredoxin fold or RNA recognition motif (RRM) found in numerous Cas proteins that are referred to as repeat-associated mysterious proteins (RAMPs) (Makarova et al. 2011b). Some RAMPs are nucleases (e.g., Cas6) (Brouns et al. 2008; Carte et al. 2008). Cmr2 is a member of the Cas10 “large subunit” Cas protein superfamily (Hale et al. 2009; Makarova et al. 2011a). Cmr2 contains an adenylyl cyclase-like domain that forms a nucleotide and metal-binding pocket (Cocozaki et al. 2012). Cmr2 also contains a predicted N-terminal HD nuclease domain, although this domain is dispensable for target RNA cleavage by the Cmr complex (Zhu and Ye 2012; Shao et al. 2013; Staals et al. 2013). Finally, Cmr5 is a member of the “small subunit” Cas protein superfamily and is primarily α-helical in structure (Sakamoto et al. 2009; Park et al. 2013; Reeks et al. 2013). The functions of the individual Cmr proteins are currently unknown.
The mechanisms involved in target RNA cleavage by Cmr complexes are a topic of ongoing investigation. Native Cmr complexes associate with multiple size forms of crRNAs and cleave complementary target RNAs at multiple sites in the region recognized by the crRNA. For example, native P. furiosus Cmr complexes contain two major crRNA species and cleave complementary target RNAs primarily at two sites (Hale et al. 2009). However, we found that Cmr complexes reconstituted with single crRNA species generated a single cleavage product at the site located 14 nt from the 3′ end of the respective crRNA species, leading to the model that each Cmr complex cleaves the target RNA a fixed distance from the 3′ end of the crRNA (Hale et al. 2009). T. thermophilus Cmr complexes contain at least three distinct crRNA species and cleave target RNAs at up to five sites (Staals et al. 2013). Studies with reconstituted T. thermophilus complexes suggest that the complex cleaves at multiple sites measured from the 5′ end of the crRNA (Staals et al. 2013). Moreover, the molecular basis of the nucleolytic activity of the Cmr complex is unknown.
We determined the basic organization and structure of the P. furiosus Cmr complex by a combination of RNA–protein cross-linking and cryo-electron microscopy (cryo-EM) of assembled Cmr complexes (Fig. 1A; Spilman et al. 2013). Our analysis revealed that Cmr4–Cmr5 units comprise the “backbone” of the particle and follow a helical path along the guide region of the crRNA. Cmr2 and Cmr3 form a “foot” at the 5′ end of the crRNA, and Cmr1 and Cmr6 form a “cap” at the 3′ end (Fig. 1A; Spilman et al. 2013). The EM structure of the T. thermophilus Cmr particle has a very similar shape, and Staals et al. (2013) developed a very similar model of the organization of the proteins in the T. thermophilus complex, while that of the Cmr complex variant characterized from S. solfataricus differs significantly (Zhang et al. 2012). Native mass spectrometric analysis of the T. thermophilus complex determined that the Cmr complex includes four Cmr4 and three Cmr5 subunits (Staals et al. 2013).
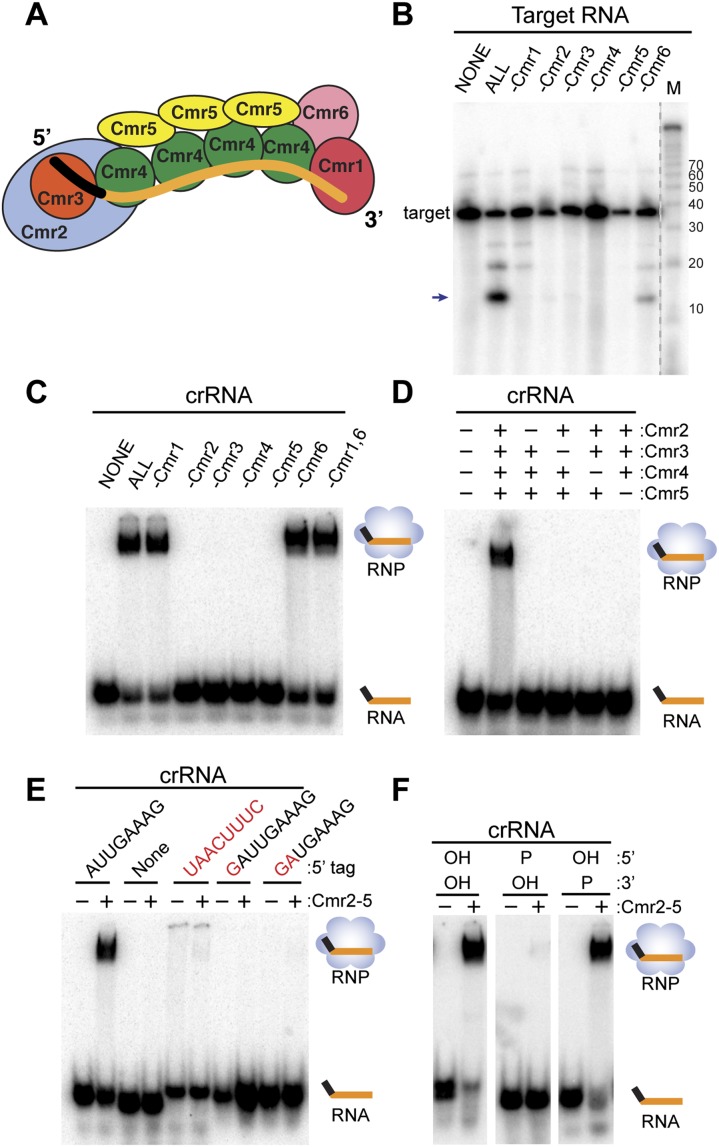
Figure 1.
Requirements for target RNA cleavage and crRNA binding by the Cmr complex. (A) Model of the organization of the Cmr protein subunits relative to the crRNA within the Cmr complex (Spilman et al. 2013). (B) Cmr protein requirements for target RNA cleavage. The indicated recombinant Cmr proteins were preincubated with the 45-nt crRNA followed by addition of 5′ end-labeled complementary target RNA. The resulting products were separated by denaturing gel electrophoresis. Radiolabeled Decade marker RNAs (M) were used for size determination. The major target RNA cleavage product is indicated (arrow). (C) Cmr protein requirements for crRNA binding. The indicated recombinant Cmr proteins were incubated with 3′ end-labeled 45-nt crRNA. RNA and complexes were resolved by native gel electrophoresis. (D) Cmr protein requirements for formation of the Cmr2–5 crRNP. The indicated Cmr proteins (Cmr2–5) were incubated with 3′ end-labeled 45-nt crRNA, and complex formation was analyzed by native gel electrophoresis. (E) 5′ tag sequence requirements for formation of the Cmr2–5 crRNP. Recombinant Cmr2–5 proteins were incubated with crRNAs that contain either a wild-type (AUUGAAAG) 5′ repeat tag, no tag, or a mutated tag sequence (complete substitution, addition of 5′ G nucleotide, or substitution of first 2 nt of 8 nt; indicated in red), and complex formation was analyzed by native gel electrophoresis. (F) End group requirements for formation of the Cmr2–5 crRNP. Recombinant Cmr2–5 proteins were incubated with crRNAs containing the indicated 5′ and 3′ chemical end groups, and complex formation was analyzed by native gel electrophoresis. Diagrams indicate the positions of unbound crRNA and crRNPs (in the absence and presence of proteins, respectively) in C–F.
In this study, we investigated the processes of Cmr complex assembly, target RNA capture, and target RNA cleavage and the roles of the Cmr proteins in these processes.
Results
All six Cmr proteins contribute to function of the type III-B effector complex
The Cmr complex from P. furiosus cleaves RNAs complementary to the guide region of the crRNA in vitro (Hale et al. 2009, 2012). In Figure 1B, we show the results of single-protein omission studies with the reconstituted complex that indicate that full activity requires all six Cmr proteins. We had previously reported cleavage activity in the absence of Cmr5 (Hale et al. 2009) but inferred that a technical issue (such as cross-contamination of a protein sample) must account for the previously observed activity. In multiple subsequent experiments, we consistently observed greatly reduced levels of activity in the absence of any individual Cmr protein in our assays (Fig. 1B; data not shown). These results indicate that efficient target RNA cleavage requires all six Cmr proteins.
Four Cmr proteins—Cmr2, Cmr3, Cmr4, and Cmr5—are required to bind a crRNA
To begin to understand the functional organization of the Cmr complex and the specific roles that each of the six Cmr proteins play, we investigated which Cmr proteins are required for recognition and incorporation of crRNAs into the complex by native gel electrophoresis with radiolabeled crRNAs (Fig. 1C). A stable crRNP assembles when all six Cmr proteins are present. Formation of a complex with the crRNA also occurs in the absence of each Cmr1 and Cmr6; however, Cmr2, Cmr3, Cmr4, and Cmr5 are each required for crRNA incorporation. In addition, Cmr2–5 can form a stable crRNP in the absence of both Cmr1 and Cmr6, and formation of the Cmr2–5 crRNP also requires the presence of all four of the individual proteins (Fig. 1D). These results indicate that four Cmr proteins—Cmr2, Cmr3, Cmr4, and Cmr5—are all essential for specific binding of a crRNA.
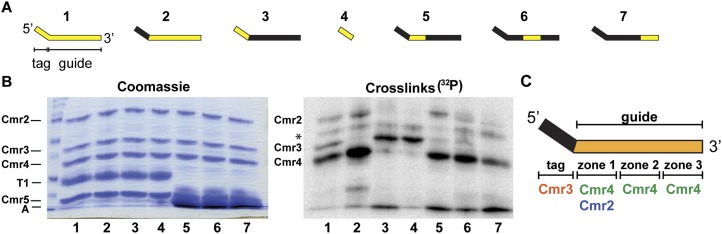
UV light-induced RNA–protein cross-linking analysis indicates that the crRNA-binding complex formed by Cmr2–5 has the same organization as the full complex formed by all six proteins, which we characterized previously (Fig. 2; Spilman et al. 2013). We performed UV cross-linking experiments on the Cmr2–5 complex with crRNAs containing 4-thiouridine (a zero-length photoactive cross-linking agent) and 32P-radiolabeled nucleotides (a means of detecting protein–RNA cross-links). The 32P-label was incorporated either throughout the entire molecule (substrate 1) or within particular regions of the crRNA, including the CRISPR-derived 5′ tag element (substrates 3 and 4), the entire guide sequence (substrate 2), or three subregions of the guide sequence (substrates 5–7) (Fig. 2A, yellow portions). The six Cmr proteins are visualized by Coomassie staining (Fig. 2B, left panel), and those covalently associated with radiolabeled regions of the crRNA are identified by autoradiography (Fig. 2B, right panel).
Figure 2.
Cmr protein–crRNA contacts in the Cmr2–5 complex. (A) UV cross-linking substrates used to analyze the locations of Cmr protein–crRNA contacts in the Cmr2–5 subcomplex. Labeled crRNAs included in preparations used for UV cross-linking assays (1–7) are diagrammed with the tag and guide regions delineated. Yellow segments indicate regions containing radiolabeled nucleotides. (B) Electrophoretic positions of Cmr proteins (left panel) and cross-linked radiolabeled nucleotides (right panel) following UV cross-linking. crRNA substrates diagrammed in A (1–7) were incubated with Cmr proteins 2–5 and exposed to UV light. The complexes were denatured and subjected to digestion by either RNase T1 or A. (Left panel) The samples were separated on SDS-PAGE gels, followed by Coomassie staining. The positions of the Cmr proteins and the RNases are indicated at the left. The crRNA substrates are indicated at the bottom of the lane. (Right panel) The Coomassie-stained gel (shown in the left panel) was subjected to phosphorimaging. The positions of the Cmr proteins with observable cross-linking to crRNAs are indicated. The asterisk indicates a nonspecific protein observed in all reactions. (C) Summary of observed Cmr protein–crRNA cross-links. The regions of the crRNA where the indicated Cmr subunits were observed to cross-link are illustrated.
Cmr2, Cmr3, and Cmr4 were cross-linked to the fully labeled crRNA, indicating that these three proteins interact directly with the crRNA in this complex (Fig. 2B, right panel, lane 1). Cmr3 was cross-linked to nucleotides in the 5′ tag element in both the context of the entire crRNA (Fig. 2B, lane 3) and the 8-nt tag RNA alone (Fig. 2B, lane 4), while Cmr2 and Cmr4 were cross-linked to nucleotides in the guide region (Fig. 2B, lane 2). Cross-linking with crRNAs labeled in three zones within the guide sequence (Fig. 2B, lanes 5–7) indicates that Cmr2 contacts the 5′-proximal region of the crRNA guide sequence (Fig. 2B, lane 5) and that Cmr4 interacts along the length of the guide sequence, with a reduction in Cmr4 signal in the 3′-terminal zone (Fig. 2B, lanes 5–7). The protein cross-linking patterns observed for the Cmr2–5 complex (summarized in Fig. 2C) mirror those observed with the crRNP assembled with the entire set of six Cmr proteins, indicating that specific interaction of Cmr2–5 with the crRNA does not depend on the Cmr1 and Cmr6 subunits. Interestingly, while Cmr5 does not appear to interact directly with the crRNA (Fig. 2; Spilman et al. 2013), it is required for association of Cmr2, Cmr3, and Cmr4 with the crRNA (Fig. 1C,D).
Formation of the Cmr complex depends on the 5′ tag and 5′ hydroxyl group of the crRNA
RNA sequencing (RNA-seq) analysis has shown that native P. furiosus Cmr complexes associate with ∼200 different crRNAs generated from the seven CRISPR loci present in this organism (Hale et al. 2012). The Cmr-associated crRNAs include an 8-nt CRISPR sequence at the 5′ end that is required for functional Cmr crRNPs (Hale et al. 2012; Zhang et al. 2012). The 5′ tag is the product of endonucleolytic cleavage at that site within the repeat sequence of CRISPR locus transcripts by Cas6 (Brouns et al. 2008; Carte et al. 2008). Cas6 cleavage generates a 5′-OH end that is also retained on the crRNAs (Carte et al. 2008; Hale et al. 2012).
Native gel mobility shift analysis indicates that formation of the Cmr complex requires both the 5′ tag sequence and 5′-OH group of the crRNA. Deletion or modification of the 5′ tag sequence disrupts binding of Cmr2–5 (Fig. 1E). Substitution of all 8 nt or of the first 2 nt of the tag sequence and even addition of a single nucleotide at the 5′ end of the tag prevent assembly (Fig. 1E). Alteration of the 5′-terminal OH group to a 5′ phosphate also prevents assembly (Fig. 1F). In contrast, modification of the end group at the 3′ terminus of the crRNA—from the 3′ phosphate found on crRNAs in vivo (Hale et al. 2012) to a 3′-OH—does not significantly affect binding by Cmr2–5 (Fig. 1F).
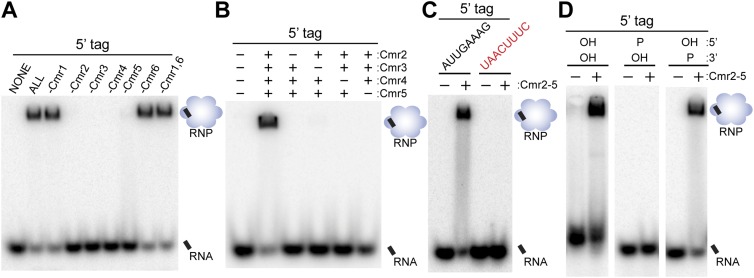
Interestingly, we found that the 5′ tag is also sufficient for crRNP formation; an RNA comprised solely of the 8-nt 5′ tag sequence forms a complex with the same characteristics as the full-length crRNA. Formation of a complex with the 5′ tag also requires all four proteins: Cmr2, Cmr3, Cmr4, and Cmr5 (Fig. 3A,B). The interaction of the Cmr proteins with the 5′ tag is also sequence-dependent and 5′-OH group-dependent (Fig. 1C,D). In addition, Cmr3 directly contacts the 5′ tag in the absence as well as the presence of the guide region of the RNA, and Cmr2, Cmr4, and Cmr5 do not contact the tag (Fig. 2B, lanes 3,4). Taken together, the results provide clear evidence for a key role of the 5′ tag sequence, including the terminal OH group, in providing specificity for Cmr protein recognition of crRNAs.
Figure 3.
Requirements for recognition of the 5′ repeat-derived crRNA tag sequence. In all panels, the indicated recombinant Cmr proteins were incubated with an 8-nt tag RNA (modified where indicated and generally 3′ end-labeled unless noted). RNA and complexes were resolved by native gel electrophoresis. Diagrams indicate the positions of unbound crRNA and crRNPs (in the absence and presence of proteins, respectively). (A) Individual Cmr proteins required for 5′ tag RNA binding by the full complex. (B) Individual Cmr proteins required for 5′ tag RNA binding by the Cmr2–5 subcomplex. (C) Sequence required for 5′ tag RNA binding by the Cmr2–5 subcomplex. Wild-type (AUUGAAAG) and mutated (UAACUUUC) tag RNAs were analyzed as indicated. (D) End groups required for 5′ tag RNA binding by the Cmr2–5 subcomplex. See the Materials and Methods for details about preparation of RNAs with the indicated 5′ and 3′ chemical end groups.
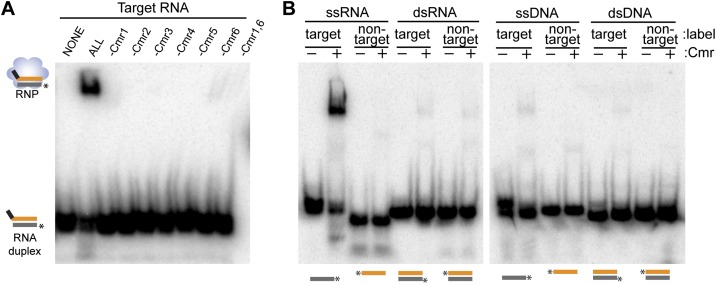
Cmr1 and Cmr6 are important for target RNA binding
We also investigated the requirements for binding the target RNA using native gel electrophoresis. (In these experiments, the target RNA is radiolabeled, and the crRNA is unlabeled.) Remarkably, we found that the Cmr2–5 crRNP is unable to efficiently bind target RNA (under conditions where crRNAs and target RNAs hybridize in the absence of proteins) (Fig. 4A, Cmr1 and Cmr6). This interesting result suggests that the crRNA is inaccessible for interaction with target RNAs within the Cmr2–5 crRNP. Indeed, both Cmr1 and Cmr6 as well as the four crRNA-binding proteins are required for target RNA capture; elimination of any of the six Cmr proteins dramatically reduces the amount of observed target RNA binding (Fig. 4A). Binding of the substrate by the Cmr complex is highly specific: No significant binding of noncomplementary ssRNA, dsRNA, ssDNA, or dsDNA, including a ssDNA complementary to the crRNA guide region, was observed (Fig. 4B).
Figure 4.
Requirements for target binding by the Cmr complex. (A) Cmr protein requirements for target RNA binding. The indicated recombinant Cmr proteins and unlabeled 45-nt crRNA were combined and incubated with 5′ end-labeled complementary target RNA. Complex formation was analyzed by native gel electrophoresis. (B) Target binding specificity. Recombinant Cmr1–6 and unlabeled 45-nt crRNA were combined and incubated with the indicated radiolabeled RNA or DNA substrates. Substrates are illustrated below each lane; asterisk indicates label location and substrate strand. “Target” substrates (gray) are complementary to the crRNA. “Nontarget” substrates (orange) are not complementary (equivalent to guide sequence of crRNA). Diagrams at the left of the panel indicate the electrophoretic positions of unbound labeled substrate (in the presence of crRNA) and crRNPs (in the presence of proteins).
Multiple cleavages in the target RNA-binding region
Native Cmr complexes contain multiple size forms of crRNAs and cleave target RNAs at multiple sites within the region of base-pairing (Hale et al. 2009, 2012; Staals et al. 2013). In initial experiments with Cmr complexes reconstituted with single crRNA species, we found a single prominent product in cleavage reactions (Hale et al. 2009, 2012); however, we observed significant additional products in some subsequent experiments, suggesting multiple cleavages. For example, in Figure 1B, in addition to the primary 14-nt product of cleavage of the 5′-labeled target RNA (Fig. 1A, arrow), smaller but significant quantities of several longer cleavage products are also observed. Similarly, T. thermophilus Cmr complexes reconstituted with a single crRNA species produce several products from a 5′-labeled target (Staals et al. 2013).
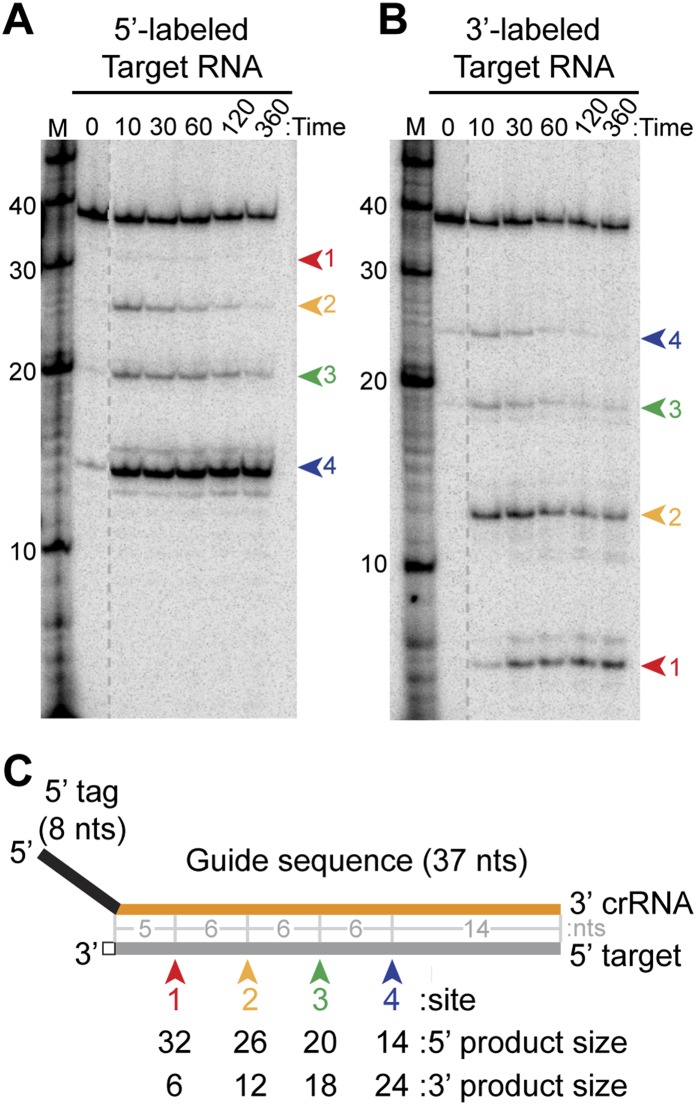
To investigate the mechanism of target RNA cleavage in detail, we analyzed 3′ end-labeled target RNAs as well as earlier time points in cleavage reactions with 5′ end-labeled target RNAs (Fig. 5). The results confirmed the presence of specific additional target RNA cleavages by P. furiosus complexes reconstituted with the 45-nt crRNA (Fig. 5). At earlier time points in the reaction, three longer 5′-labeled cleavage products are more readily detected, and with 3′-labeled target RNA substrates, corresponding cleavage products are observed (Fig. 5A,B, red, yellow, and green arrows at sites 1, 2, and 3). The additional target RNA cleavages occur at 6-nt intervals from the originally detected site (Fig. 5A,B, blue arrow at site 4), which is located 14 nt from the 3′ end of the paired crRNA (Fig. 5A,B [and additional nucleotide-resolution mapping studies], C; data not shown). The smallest product detected in cleavage of the 3′-labeled target RNA corresponds to cleavage at site 1 (Fig. 5B,C, red arrow), 5 nt from the 5′ tag of the crRNA (Fig. 5B,C). Multiple cleavage products can also be observed with complexes reconstituted with the other major Cmr crRNA species: Complexes reconstituted with the 39-nt crRNA cleave at sites 1 and 2 (Fig. 6A,B, red and yellow arrows) in addition to the primary cleavage originally observed at site 3 (Fig. 6A,B, green arrow, “39” lane). Thus, Cmr complexes reconstituted with single crRNA species cleave target RNAs at regular 6-nt intervals within the region of complementarity: four cleavages with the 45-nt crRNA and three cleavages with the 39-nt crRNA. The 39- and 45-nt crRNAs guide cleavage at the same sites on the target RNA (with the exception of site 4) (Fig. 6A,B).
Figure 5.
Analysis of cleavage of 5′- and 3′-labeled target RNAs. (A,B) Cleavage of 5′- and 3′-labeled target RNAs by the Cmr complex. Recombinant Cmr1–6 proteins and unlabeled 45-nt crRNA were combined and incubated with 5′-labeled (A) or 3′-labeled (B) complementary target RNAs. Samples were incubated for the indicated amount of time at 70°C or for 360 min on ice (ice), and the labeled target RNAs were separated by high-resolution denaturing polyacrylamide gel electrophoresis. Colored arrows indicate the products of cleavage at the four observed cleavage sites. Radiolabeled Decade marker RNAs (M) were used for size determination. The dotted line indicates noncontiguous lanes. (C) Summary of the mapped target RNA cleavage sites. The target RNA (gray) is fully complementary to the guide region (orange) of the crRNA used in the assay. Colored arrows indicate the positions of the observed cleavages, corresponding to the products observed in A and B. The sizes of the products observed in reactions with 5′- and 3′-labeled substrates are indicated below, and distances between observed cleavage sites are shown in gray. The 3′ end-labeled target RNA is generated by ligation of a radiolabeled pCp moiety, which adds a single nucleotide to the 3′ end of the target RNA (indicated with a box).
Figure 6.
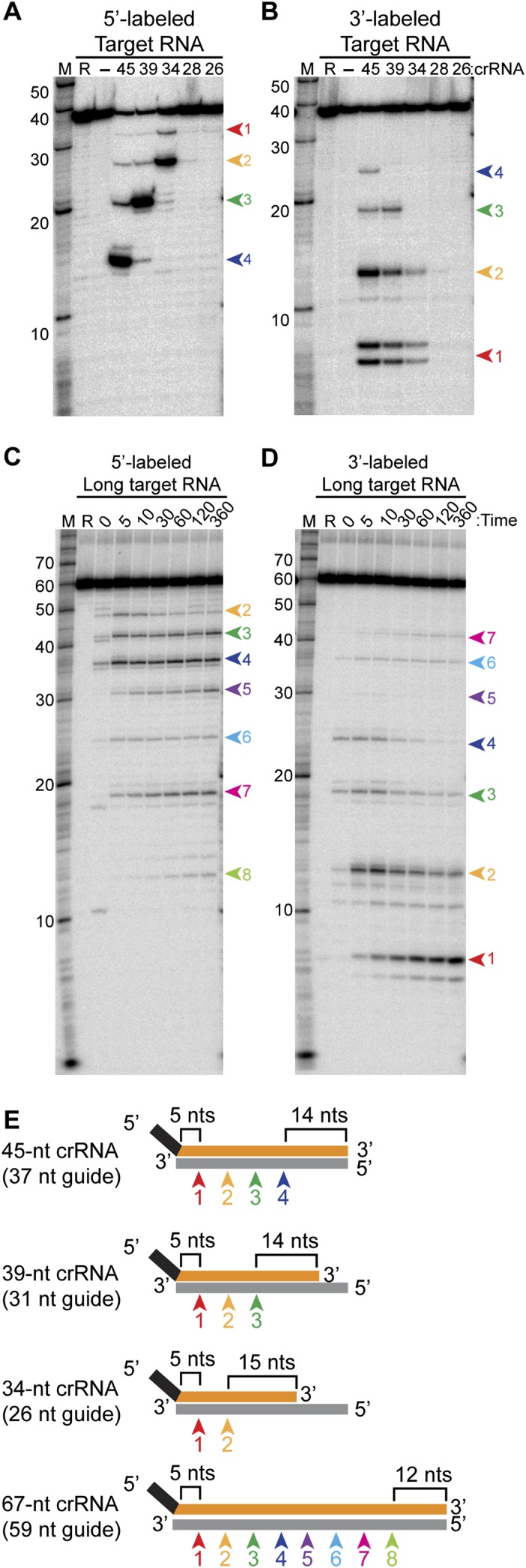
Analysis of cleavages directed by crRNAs of variable guide sequence length. (A,B) Cleavage directed by Cmr complexes containing crRNAs with a series of shorter guide sequences. Recombinant Cmr1–6 and unlabeled crRNAs of the indicated lengths were combined and incubated with 5′ end-labeled (A) or 3′ end-labeled (B) complementary target RNA. (R) Target RNA alone; (−) no crRNA was added. Cleavage products were separated by high-resolution denaturing polyacrylamide gel electrophoresis. Colored arrows indicate the products of cleavage at the four observed cleavage sites (labeled as in Fig. 5). The doublet observed for the smallest product of the 3′-labeled target RNA (cleavage site 1) is likely due to partial loss of the 3′ phosphate added during the 3′ pCp labeling reaction. (B) Cleavages by Cmr complexes containing a crRNA with an extended guide sequence. Recombinant Cmr1–6 and unlabeled 67-nt crRNAs were combined and incubated with 5′ end-labeled (C) or 3′ end-labeled (D) complementary target RNA. The reactions were incubated for the indicated amount of time at 70°C or for 360 minutes on ice (ice). (Target RNA alone [R] was incubated for 360 minutes at 70°C.) Colored arrows indicate the products of cleavage at the eight observed cleavage sites. In A–D, radiolabeled Decade marker RNAs (M) were used for size determination. (E) Summary of the mapped target RNA cleavage sites. Sites of cleavage guided by crRNAs of various lengths are indicated by colored arrows and correspond to the products observed in A–D. The length of the crRNA guide region is indicated in parentheses, and the distances from the ends of the guide region to the nearest site of cleavage are indicated.
Our data suggest that each target RNA molecule is cleaved at multiple sites in these reactions. While the major product of cleavage of the 5′-labeled target RNA by the 45-nt crRNA complex corresponds to cleavage at site 4 (nearest the 3′ end of the crRNA), the primary products observed with the 3′-labeled substrate correspond to cleavage at sites 1 and 2 (nearest the 5′ end of the crRNA) (Figs. 5, 6A,C). The site 4 cleavage product is the predominant product observed with the 5′-labeled target even at the earliest time points examined, and levels of the longer, less abundant products decrease over time, presumably as a result of their further cleavage (Fig. 5). Similarly, the site 2 cleavage product is the major product detected at the earliest time point with the 3′-labeled target, and longer products decrease over time (Fig. 5), suggesting that the target RNAs have already been cleaved multiple times at the early (10-min) reaction time point. Interestingly, cleavage at site 1 (proximal to the crRNA tag) appears to be delayed relative to the other sites (Fig. 5A, red arrow). The results suggest that the cleavages at sites 2–4 (11, 17, and 23 nt from the 5′ tag of the crRNA) are favored over cleavage at site 1 by the reconstituted Cmr complexes.
Parameters of cleavage site targeting by Cmr crRNAs
Because the cleavage detected in our early studies with Cmr complexes reconstituted with each the 39-nt and 45-nt crRNA was located 14 nt from the 3′ end of the respective crRNA, we hypothesized that crRNA-guided cleavage occurs a fixed distance (14 nt) from the 3′ end of the crRNA (Hale et al. 2009). However, the additional cleavages now observed also begin a fixed distance from the 5′ end of the crRNA (5 nt from the 5′ tag sequence).
To better understand the mechanism of cleavage site targeting by crRNAs in P. furiosus Cmr complexes, we tested the cleavage patterns generated by crRNAs of various lengths (Fig. 6A–D). The efficiency of target RNA cleavage decreased when the length of the guide sequence was reduced to 20 nt, presumably due to reduced target RNA hybridization and/or less efficient complex formation (Fig. 6A,B, “28” and “26” crRNA lanes). Overall, as the crRNA guide sequence is shortened from the 3′ end, the cleavages at the corresponding 5′ end of the target RNA are progressively lost (Fig. 6A,B,E). On the other hand, the number of cleavages achieved by the Cmr complex can be increased by elongating the guide sequence: Increasing the length of the guide region of the 45-nt crRNA by 22 nt led to four additional target RNA cleavages at 6-nt intervals (Fig. 6C–E, 67-nt crRNA). Importantly, however, the locations of the cleavages shifted relative to the 3′ ends of the crRNAs but remained constant relative to the 5′ ends of the various crRNAs. The distance between the 3′ end of the crRNA and the nearest site of cleavage was 15 nt for the 34-nt crRNA and 12 nt for the 67-nt crRNA (Fig. 6A,C). The results indicate that cleavage sites are determined by distance from the 5′ (tag) end of the crRNA rather than from the 3′ end of the crRNA, as we had previously hypothesized (Hale et al. 2009).
Together, our analyses reveal that P. furiosus Cmr complexes can cleave complementary target RNAs at sites located at 6-nt intervals along the length of the target–crRNA duplex beginning 5 nt downstream from the crRNA tag. Cleavages can occur up through a region of ∼14 nt at the end of the crRNA–target RNA duplex; no substantial cleavage is observed at other potential sites in this region (6 and 12 nt downstream from the last observed cleavage).
Discussion
The type III-B Cmr immune effector complex is a multisubunit noncoding RNP that carries out RNA-guided cleavage of invader target RNAs in diverse bacterial and archaeal organisms (Haft et al. 2005; Hale et al. 2009, 2012; Garrett et al. 2011; Makarova et al. 2011b; Zhang et al. 2012; Bailey 2013; Staals et al. 2013). In this study, we determined how the organization of the RNP relates to functions, including crRNA loading, target RNA capture, and target RNA cleavage (Fig. 7).
Figure 7.
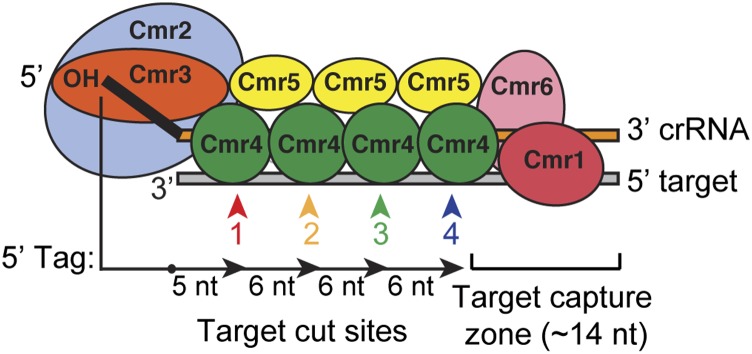
Functional organization of the Cmr complex. The organization of the Cmr proteins along the length of the crRNA in the P. furiosus Cmr complex is shown (Spilman et al. 2013). The findings presented here provide significant functional insights on this complex. Four of the Cmr proteins—Cmr2–5 (blue, orange, green, and yellow)—are required to specifically bind RNAs with the signature CRISPR tag sequence (Fig. 1); Cmr3 interacts with the tag directly (Fig. 2; Spilman et al. 2013). Target RNA capture additionally requires Cmr1 and Cmr6 (red and pink) (Fig. 4). Cmr complexes cleave target RNAs at multiple sites located at 6-nt intervals in the region of complementarity, beginning 5 nt downstream from the crRNA tag (Figs. 5, 6). Cleavages can occur up through a region of ∼14 nt at the end of the target RNA–crRNA duplex. This target capture zone is critical for target RNA cleavage (Hale et al. 2009) and is the binding site of Cmr1 and Cmr6 (Spilman et al. 2013), which are critical for target RNA capture (Fig. 4). Our data indicate that Cmr4 plays a critical role in specifying the target RNA cleavages and may be the catalytic component of the Cmr complex.
crRNA binding requires four Cmr proteins
We found that within the multisubunit Cmr complex, four proteins are involved in crRNA binding. Cmr3 cross-links with the signature 5′ tag sequence of the crRNAs (Fig. 3; Spilman et al. 2013). The 5′ tag is the defining feature of the crRNAs, is essential for Cmr complex formation and function (Figs. 1–3), and can also be used to engineer functional crRNAs to direct cleavage of novel target RNAs by the Cmr complex (Hale et al. 2012). Interestingly, however, the interaction of Cmr3 with crRNAs requires three additional proteins: Cmr2, Cmr4, and Cmr5 (Figs. 1, 3). Cmr2 interacts with Cmr3 (Osawa et al. 2013; Shao et al. 2013; Spilman et al. 2013; Staals et al. 2013) and with the guide sequence of the crRNA nearest the 5′ tag (Fig. 2; Spilman et al. 2013). Another recent study found that the Cmr2–Cmr3 complex could bind the crRNA and target RNA in the absence of the other Cmr proteins (Osawa et al. 2013); however, we did not observe specific RNA binding by these two proteins under the conditions used in our work (with competitor RNA to reduce nonspecific interactions). Four Cmr4 subunits form a helical chain (Staals et al. 2013; N Ramia, M Spilman, L Tang, Y Shao, J Elmore, CR Hale, A Cocozaki, N Bhattacharya, RM Terns, MP Terns, et al., unpubl.) along the majority of the length of the guide region of the crRNA in both the minimal crRNP (Cmr2–5) and the full complex (Spilman et al. 2013). Three Cmr5 subunits form a protein chain parallel to the Cmr4 chain (Spilman et al. 2013; Staals et al. 2013) and do not appear to directly interact with the crRNA (Fig. 2; Spilman et al. 2013) but nonetheless are essential for the interaction of the other three proteins—Cmr2, Cmr3, and Cmr4—with the crRNA (Figs. 1, 3).
Our findings indicate that Cmr3 recognizes the signature crRNA sequence motif but requires protein–protein interactions within a complex that also includes Cmr2, Cmr4, and Cmr5 for full crRNA-binding activity. Cmr2–5 form a complex with a crRNA interaction pattern and tag sequence dependence that are indistinguishable from the full complex (Figs. 1, 3). Each of the four proteins is required for formation of the complex (Fig. 1). However, the proteins also interact with an RNA comprised of only the 5′ tag sequence (lacking the guide region that is bound by Cmr2 and Cmr4) (Fig. 3), and all four proteins are still essential for crRNA binding. This finding indicates that Cmr2 and Cmr4 do not simply stabilize interaction of Cmr3 with the crRNA by binding the guide region but that protein–protein interactions among the Cmr2, Cmr3, Cmr4, and Cmr5 proteins are needed for Cmr3 to specifically recognize and bind crRNAs.
Cmr recognition of target RNA
Remarkably, efficient target RNA capture depends on the additional presence of Cmr1 and Cmr6. Despite the presence of the crRNA, which includes a 37-nt guide region that is complementary to the target RNA, the crRNP formed by Cmr2–5 does not substantially interact with the target RNA in our assays (Fig. 4). Under the same conditions, the crRNA interacts with the target RNA in the absence of proteins (Fig. 4). Our findings suggest that the crRNA guide region is unavailable for target RNA interaction in the Cmr2–5 crRNP and that Cmr1 and Cmr6 are required to induce an accessible conformation and facilitate target RNA capture.
Both Cmr1 and Cmr6 are RAMPs with predicted RRMs (Makarova et al. 2011a). Cmr1 and Cmr6 form the cap of the Cmr complex (at the opposite end of the Cmr4–Cmr5 helix from Cmr2 and Cmr3) and contact the 3′ end of the crRNA (Spilman et al. 2013). In previous studies, we found that complementarity of the target to the 3′ end of the crRNA (where Cmr1 and Cmr6 are located) is essential for target RNA cleavage (Hale et al. 2009). While site 4 cleavage was unaffected by the absence of target sequences complementary to nearly the first half of the guide region of the 45-nt crRNA (lacking 18 nt of the 37-nt guide region), cleavage was not observed in the absence of the target sequence complementary to the last 6 nt at the 3′ end of the crRNA (Hale et al. 2009). Our work indicates that Cmr1 and Cmr6 mediate a critical interaction with the substrate at the 3′ end of the crRNA. The molecular mechanism by which Cmr1 and Cmr6 enable target RNA capture may be through protein–protein interaction (e.g., Cmr6 interacts with Cmr4 [Park et al. 2013; CR Hale, RM Terns, and MP Terns, unpubl.]) and/or direct binding to the 3′ end of the crRNA (Cmr1 and Cmr6 both cross-link with the 3′ region of the crRNA [Spilman et al. 2013]). It is not yet known whether Cmr1 and Cmr6 also interact directly with the target RNA to facilitate capture.
crRNA 5′ tags and Cas5 proteins in type I and type III CRISPR–Cas systems
crRNAs from all known type I and type III CRISPR–Cas systems have 5′ CRISPR tags. Work establishing the importance of 5′ tags in the type III-B Cmr system (Hale et al. 2009, 2012; Zhang et al. 2012) supports the broad importance of crRNA tags in crRNP formation and function. Interestingly, our study also revealed a critical role for the 5′-OH group of the tag in the assembly of Cmr crRNPs (Figs. 1, 3). The 5′ end group of eukaryotic siRNAs (phosphate in this case) is also vital for RNP assembly and function (Elbashir et al. 2001; Ma et al. 2004; Wang et al. 2009). Many processed RNAs found in prokaryotic cells have 5′ phosphate ends, but crRNAs acquire 5′-OH termini as the result of Cas6 cleavage of CRISPR transcripts (Brouns et al. 2008; Carte et al. 2008; Hale et al. 2012). The 5′-OH ends of mature crRNAs contribute to successful discrimination against other RNAs whose incorporation into CRISPR–Cas effector complexes would be detrimental to the cell.
The structural organization of the subunits in the Cmr complex is strikingly mirrored by that of distantly related superfamily members in complexes from other type I and type III CRISPR–Cas systems (Wiedenheft et al. 2011a; Rouillon et al. 2013; Spilman et al. 2013; Staals et al. 2013; Jackson et al. 2014; Mulepati et al. 2014; Zhao et al. 2014), and our findings predict potential roles for those related subunits. In particular, Cmr3, the protein that our results implicate in recognition of the 5′ tag, is a member of the diverse Cas5 superfamily (Makarova et al. 2011b). Members of the Cas5 superfamily are found in type I and type III CRISPR–Cas systems. (type II CRISPR–Cas systems lack Cas5 homologs, and the mature crRNAs lack 5′ tags [Deltcheva et al. 2011; Makarova et al. 2011b]). Our finding that Cmr3 directly contacts the 5′ tag (Fig. 2; Spilman et al. 2013) and is essential for crRNA recognition (Figs. 1, 3) provides evidence that Cas5 superfamily proteins play an important and conserved role in crRNA recognition and binding in type I and type III CRISPR–Cas systems. Indeed, very recent X-ray structures of type I-E crRNPs reveal intimate and base-specific contacts between the 5′ tag of the crRNA and residues of the Cas5 homolog of this system (Jackson et al. 2014; Mulepati et al. 2014; Zhao et al. 2014).
Evidence for multiple cleavages mediated by Cmr4
Reconstituted P. furiosus Cmr complexes include a helical chain of four Cmr4 subunits arrayed along the length of the crRNA guide region (Spilman et al. 2013; N Ramia, M Spilman, L Tang, Y Shao, J Elmore, CR Hale, A Cocozaki, N Bhattacharya, RM Terns, MP Terns, et al., unpubl.). The Cmr4 arrangement in the P. furiosus complex correlates nicely with the four evenly spaced cleavages observed on target RNAs (Figs. 5, 6), suggesting that cleavages occur at each Cmr4 subunit (Fig. 7). The dimensions of the Cmr4 protein structure correspond well with the 6-nt cleavage site interval between cleavage sites (Spilman et al. 2013; Staals et al. 2013). In addition, we found that mutation of P. furiosus Cmr4 (D26 to N) abolishes cleavage activity without disrupting formation of a crRNA-containing complex (N Ramia, M Spilman, L Tang, Y Shao, J Elmore, CR Hale, A Cocozaki, N Bhattacharya, RM Terns, MP Terns, et al., unpubl.)
Of note, however, native T. thermophilus Cmr complexes, which have also been found to have four Cmr4 subunits, catalyze five cleavages (Staals et al. 2013). A supplemental nucleolytic subunit (e.g., Cmr1 or Cmr6) may cleave at the fifth site in Tth, but this subunit apparently does not generate a cleavage in the analogous P. furiosus complex. Alternatively, variability in Cmr complex composition could account for additional cleavages, as described below.
Cse4, the Cas7 superfamily member that forms the elongated core of the type I-E Cse complex found in E. coli, also appears to be involved in target cleavage (Jore et al. 2011; Wiedenheft et al. 2011a; Spilman et al. 2013; Staals et al. 2013; Hochstrasser et al. 2014). In the Cse complex, Cse4 subunits contact 1-nt unpaired pinch points that form between 5-nt regions of the crRNA–target DNA duplex (Wiedenheft et al. 2011a; Jackson et al. 2014; Mulepati et al. 2014; Zhao et al. 2014). By analogy, similar discontinuous pairing between the crRNA and target RNA in the Cmr complex may define the sites of cleavage at each of the Cmr4 backbone subunits. Interestingly, we found that the P. furiosus Cmr complex displays a distinct preference for cleavage at three of the four observed cleavage sites (sites 2–4) (Figs. 5, 6), suggesting a possible role for the three Cmr5 subunits that are found in parallel with the Cmr4 subunits in the Cmr complex in promoting cleavage.
Target RNA capture zone at the 3′ end of crRNA
Our findings indicate that there is a region of the target RNA associated with the 3′ end of the crRNA where cleavages do not occur (see Fig. 7). Target RNA cleavages occur at regular 6-nt intervals within the region of complementarity with the crRNA, up through the region of ∼14 nt base-paired with the 3′ end of the crRNA (Figs. 5, 6; Hale et al. 2009, 2012). For the naturally occurring crRNA size species (45- and 39-nt crRNAs), the last observed cleavage occurs 14 nt from the 3′ end of the crRNA; no substantial cleavage is observed at the next potential sites 6 and 12 nt downstream. With variant crRNAs, we found that the uncleaved region can be reduced to 12 nt (Fig. 6, 67-nt crRNA) but not to 9 nt: Cleavage is not observed at site 3 with the 34-nt crRNA (which would result in a void region of 9 nt), and instead the void region is extended to 15 nt (Fig. 6).
The observed cleavage void could reflect a requirement of the Cmr complex for a base-paired region of at least 9 nt at the 3′ end of the crRNA. We also found that cleavage activity is disrupted when the target RNA lacks the region complementary to the last 6 nt of the crRNA (Hale et al. 2009). The region of base-pairing at the 3′ end of the crRNA may be required for binding of Cmr1 and/or Cmr6–Cas7 superfamily members that do not appear to mediate cleavages in the P. furiosus Cmr complex. Indeed, Cmr1 and Cmr6 are essential for target RNA binding by the Cmr complex (Fig. 4). We propose that the 3′ end of the crRNA is a critical zone for target RNA capture requiring Cmr1 and Cmr6 (Fig. 7).
Modular assembly of Cmr complexes with variable numbers of Cmr4 subunits
P. furiosus Cmr complexes reconstituted with crRNAs of varying lengths cleave targets at different numbers of sites corresponding with the length of the crRNA guide region: from two sites with a 34-nt crRNA to eight sites with a 67-nt crRNA (Fig. 6). It is possible that complexes containing four catalytic Cmr4 subunits mediate fewer cleavages when loaded with shorter crRNAs and perhaps thread the target RNA through a nuclease site to produce additional cleavages with longer crRNAs. However, another possibility is the existence of populations of Cmr complexes with variable numbers of Cmr4 subunits—populations in addition to the particle with four Cmr4 subunits found in both reconstituted P. furiosus and native T. thermophilus samples.
We observed that Cmr4 and Cmr5 have the ability to form helical filaments of varying lengths in the absence of crRNA and the other subunits (Spilman et al. 2013), consistent with the possibility of the assembly of Cmr complexes with variable numbers of Cmr4 (and Cmr5) subunits. Moreover, the formation of particles of various sizes could also elegantly account for the existence of the series of Cmr crRNA species found in vivo (e.g., in P. furiosus [Hale et al. 2009, 2012] and T. thermophilus [Staals et al. 2013]). Notably, the interval in the sizes of native Cmr crRNAs and in the distance between sites of cleavage by Cmr complexes is the same (6 nt), strongly suggesting that units of Cmr4 could account for both. The sizes of the Cmr-associated crRNAs observed in vivo likely reflect a “footprint” of the structural nature of the Cmr complexes in a given cell. In addition, Cmr4 is a member of the diverse Cas7 superfamily of proteins, and evidence suggests that the number of subunits of Csm3, another member of this superfamily, functions in determination of the sizes of crRNAs (with 6-nt intervals) in a type III-A Csm system (Hatoum-Aslan et al. 2013).
Taken together, the simple clear model that emerges from our findings is that Cmr complexes assemble with varying numbers of Cmr4 subunits occupying 6-nt intervals along the crRNA guide region. Exonucleolytic processing of the 3′ ends of crRNAs assembled in the various forms of the Cmr complex give rise to the 6-nt ladders of crRNAs observed in vivo (with a preference for complexes assembled along the full length of the crRNA guide sequence, corresponding to complexes with four Cmr4 subunits and ∼45-nt crRNAs in vivo). Each Cmr4 subunit in the assembled complex can mediate a cleavage, giving rise to 6-nt ladders of cleavage products. (Cleavage at the first Cmr4 subunit may be compromised by its lack of a Cmr5 partner protein.) The possible existence of modularly assembled variations of the Cmr complex provides an alternative to cleavage by Cmr1 or Cmr6 to account for the fifth cleavage observed in T. thermophilus (Staals et al. 2013).
Materials and methods
Recombinant Cmr proteins
Expression and purification of recombinant Cmr proteins was performed as described (Hale et al. 2009) with some modifications. Briefly, the proteins were expressed from 1–4 L of either Luria broth (Cmr1, Cmr2dHD, Cmr4, and Cmr5) or Terrific broth (Cmr3 and Cmr6). After sonication and thermal precipitation for 30 min at 70°C, proteins were further purified on 1.5-mL columns containing either Ni-NTA (Cmr1, Cmr2dHD, Cmr3, Cmr4 and Cmr5; Qiagen) or Talon cobalt resin (Cmr6; Clontech). After dialysis (40 mM Hepes at pH 7.0, 500 mM KCl), the concentrations were determined by Qubit protein concentration assays (Life Technologies) and confirmed by SDS-PAGE and Coomassie staining.
Generation of RNA substrates
Sequences of RNA and DNA oligos used in this study can be found in Supplemental Table 1. 5′ and 3′ end 32P-labeled RNAs were generated as described previously (Carte et al. 2010). Internally labeled RNAs (Figs. 1E,F, 3D) were created by in vitro transcription (IVT) using T7 RNA polymerase (Life Technologies) for 2 h at 37°C in 20 μL reactions as recommended by the manufacturer, including 4 μL of α-[32P]-GTP (800 Ci/mmol, 10 mCi/mL; Perkin Elmer). Transcribed RNAs were gel-purified and, for RNAs in Figure 1E, subjected to either phosphatase treatment (to generate RNAs with 5′-OH groups) as described previously (Hale et al. 2009) or Cas6 digestion to generate 5′-tagged crRNAs (Figs. 1F, 3D). Cas6 cleavage reactions were performed using reaction conditions described previously (Carte et al. 2008) using 1 μM recombinant P. furiosus Cas6 in 100-μL reactions for 3 h at 70°C. The sequences of the DNA templates used for IVT reactions are provided in Supplemental Table 1. Template annealing was performed as described (Hale et al. 2009).
crRNA-binding assays
For crRNA-binding assays, the indicated recombinant Cmr proteins (500 nM) were incubated with 5000 counts per minute (cpm) (∼0.05 pmol) of 32P-radiolabeled crRNA substrates for 30 min at 70°C in 20 mM Hepes (pH 7.0), 250 mM KCl, 10% glycerol, 1.5 mM MgCl2, and 5 mM DTT supplemented with 5 μg of E. coli tRNA. Half of the reactions were added directly to nondenaturing minigels containing 0.5× TBE (44.5 mM Tris at pH 8.3, 44.5 mM boric acid, 1 mM EDTA), 6% acrylamide (acrylamide:bis acrylamide 37.5:1; Bio-Rad), and 4% glycerol. Gels were run at 110 V for 70 min, dried, and subjected to phosphorimaging.
UV cross-linking
Preparation of 4-thiouridine and 32P-labeled crRNA cross-linking substrates and UV cross-linking assays was performed as described (Spilman et al. 2013). Briefly, RNA substrates were incubated with the Cmr proteins for 30 min at 70°C. A portion of the reactions was assessed for crRNA binding by native gel electrophoresis (data not shown). The remaining reactions were subjected to UV cross-linking (312 nm) for 30 min. The cross-linked samples were denatured with 1% SDS and 70°C heating, digested with either RNase T1 (Fig. 2, substrates 1–4) or RNase A (Fig. 2, substrates 5–8), and separated on 10% SDS-PAGE gels. The resulting gel was visualized by R-250 Coomassie staining, dried, and subjected to phosphorimaging and autoradiography to allow for alignment of protein bands with autoradiographs.
Target RNA-binding and cleavage assays
Target RNA-binding assays were performed as described for crRNA binding above, with the following exception. Reactions were first assembled with 0.5 pmol of synthetic 7.01 crRNA for 30 min at 70°C prior to the addition of 5000 cpm (∼0.05 fmol) of the radiolabeled target RNA or DNA. The samples were then incubated for 30 min at 70°C and separated by electrophoresis on 6% native polyacrylamide gels. To assay for target RNA cleavage, the reactions (lacking E. coli tRNA) were subjected to phenol extraction and ethanol precipitation and run on 15% mini 1× TBE-urea (7 M) gels (Fig. 1) or sequencing-sized gels. 5′ end-radiolabeled RNAs (Decade marker; Life Technologies) were used for size standards on all denaturing gels.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants RO1GM54682 (to M.P.T. and R.M.T.) and RO1GM99604 (to H.L.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.250712.114.
References
- Bailey S. 2013. The Cmr complex: an RNA-guided endoribonuclease. Biochem Soc Trans 41: 1464–1467 [DOI] [PubMed] [Google Scholar]
- Barrangou R, Marraffini LA. 2014. CRISPR–Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell 54: 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151: 2551–2561 [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. 2008. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22: 3489–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. 2010. Binding and cleavage of CRISPR RNA by Cas6. RNA 16: 2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocozaki AI, Ramia NF, Shao Y, Hale CR, Terns RM, Terns MP, Li H. 2012. Structure of the Cmr2 subunit of the CRISPR–Cas RNA silencing complex. Structure 20: 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471: 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. 2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 20: 6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468: 67–71 [DOI] [PubMed] [Google Scholar]
- Garrett RA, Vestergaard G, Shah SA. 2011. Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol 19: 549–556 [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. 2012. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci 109: E2579–E2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP. 2008. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA 14: 2572–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. 2009. RNA-guided RNA cleavage by a CRISPR RNA–Cas protein complex. Cell 139: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV 3rd, Graveley BR, Terns RM, et al. 2012. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell 45: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Samai P, Maniv I, Jiang W, Marraffini LA. 2013. A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J Biol Chem 288: 27888–27897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. 2014. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci 111: 6618–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Nam KH, Ding F, Lee H, Wu L, Xiao Y, Farchione MD Jr, Zhou S, Rajashankar K, Kurinov I, et al. 2014. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol 21: 771–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, Brouns SJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. 2014. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 345: 1473–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. 2011. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 18: 529–536 [DOI] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ. 2004. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429: 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV. 2011a. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR–Cas systems. Biol Direct 6: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. 2011b. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9: 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322: 1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen AM, Horvath P, Boyaval P, Romero DA. 2012. Mobile CRISPR/Cas-mediated bacteriophage resistance in Lactococcus lactis. PLoS ONE 7: e51663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60: 174–182 [DOI] [PubMed] [Google Scholar]
- Mulepati S, Bailey S. 2013. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem 288: 22184–22192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Heroux A, Bailey S. 2014. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science 345: 1479–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Inanaga H, Numata T. 2013. Crystal structure of the Cmr2–Cmr3 subcomplex in the CRISPR–Cas RNA silencing effector complex. J Mol Biol 425: 3811–3823 [DOI] [PubMed] [Google Scholar]
- Park JH, Sun J, Park SY, Hwang HJ, Park MY, Shin M, Kim JS. 2013. Crystal structure of Cmr5 from Pyrococcus furiosus and its functional implications. FEBS Lett 587: 562–568 [DOI] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151: 653–663 [DOI] [PubMed] [Google Scholar]
- Reeks J, Graham S, Anderson L, Liu H, White MF, Naismith JH. 2013. Structure of the archaeal Cascade subunit Csa5: relating the small subunits of CRISPR effector complexes. RNA Biol 10: 762–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C, Zhou M, Zhang J, Politis A, Beilsten-Edmands V, Cannone G, Graham S, Robinson CV, Spagnolo L, White MF. 2013. Structure of the CRISPR interference complex CSM reveals key similarities with Cascade. Mol Cell 52: 124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Agari Y, Agari K, Yokoyama S, Kuramitsu S, Shinkai A. 2009. X-ray crystal structure of a CRISPR-associated RAMP superfamily protein, Cmr5, from Thermus thermophilus HB8. Proteins 75: 528–532 [DOI] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, Doudna JA. 2012. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell 46: 606–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Cocozaki AI, Ramia NF, Terns RM, Terns MP, Li H. 2013. Structure of the cmr2-cmr3 subcomplex of the cmr RNA silencing complex. Structure 21: 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. 2011. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 30: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. 2013. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J 32: 385–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Lawrence CM, Wiedenheft B. 2013. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem 82: 237–266 [DOI] [PubMed] [Google Scholar]
- Spilman M, Cocozaki A, Hale C, Shao Y, Ramia N, Terns R, Terns M, Li H, Stagg S. 2013. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell 52: 146–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Agari Y, Maki-Yonekura S, Zhu Y, Taylor DW, van Duijn E, Barendregt A, Vlot M, Koehorst JJ, Sakamoto K, et al. 2013. Structure and activity of the RNA-targeting type III-B CRISPR–Cas complex of Thermus thermophilus. Mol Cell 52: 135–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Terns RM. 2011. CRISPR-based adaptive immune systems. Curr Opin Microbiol 14: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B. 2014. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. 2009. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461: 754–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. 2012. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. 2011a. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477: 486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, et al. 2011b. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci 108: 10092–10097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. 2012. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell 45: 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, Wang M, Bunkoczi G, Gong W, Wei Z, Wang Y. 2014. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature doi: 10.1038/nature13733 [DOI] [PubMed] [Google Scholar]
- Zhu X, Ye K. 2012. Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR–Cas systems. FEBS Lett 586: 939–945 [DOI] [PubMed] [Google Scholar]