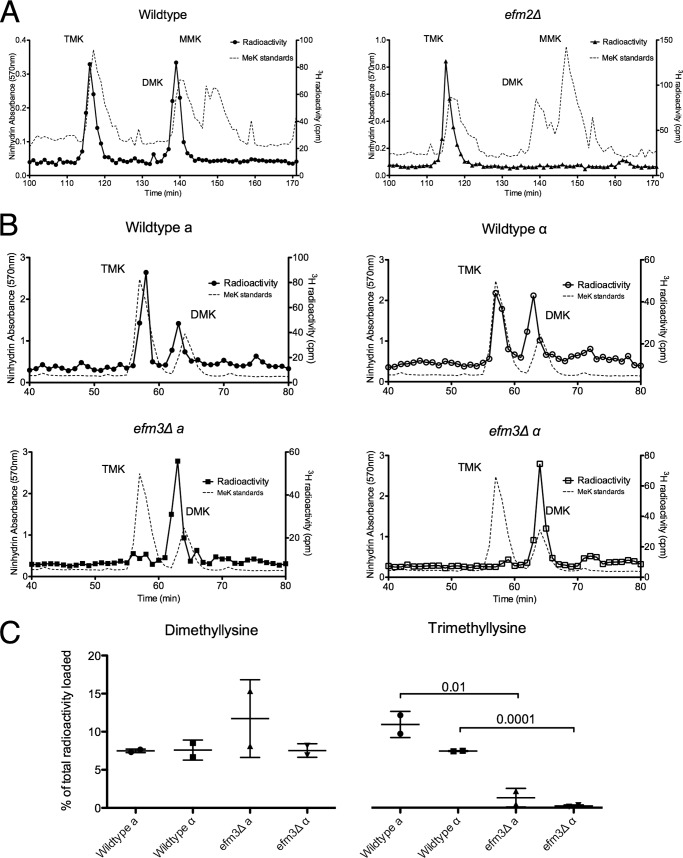
FIGURE 2.
Deletion of EFM2 results in loss of dimethylated lysine, and deletion of EFM3 results in loss of trimethylated lysine in 100-kDa polypeptides. Lysates from in vivo radiolabeled cells were fractionated by SDS-PAGE, and the 100 kDa gel region was excised. Gel slices were acid-hydrolyzed as described under “Experimental Procedures.” The resulting hydrolysates were loaded onto a high resolution cation exchange column with standards of methylated lysine derivatives. A, using a pH 3.8 elution buffer, radiolabeled methylated lysine derivatives were separated. The position of the standards, detected by ninhydrin reactivity, is shown by the dashed line. Due to a tritium isotope effect, the radiolabeled derivatives elute slightly before the non-labeled standards (58). Each trace is representative of three independent experiments. B, separation of hydrolysates from radiolabeled wild type and efm3Δ cells in both mating type backgrounds. A pH 4.5 elution buffer was used. The ninhydrin profiles are shown for methylated standards run in a separate experiment due to interference from the large amounts of ammonium ion present in the gel hydrolysates. Each trace is representative of two independent experiments. C, the amount of di- and trimethyllysine radioactivity as a percentage of the total radioactivity in the hydrolysate is shown, with error bars reflecting the S.D. p values from Student's t test are shown when less than 0.05.