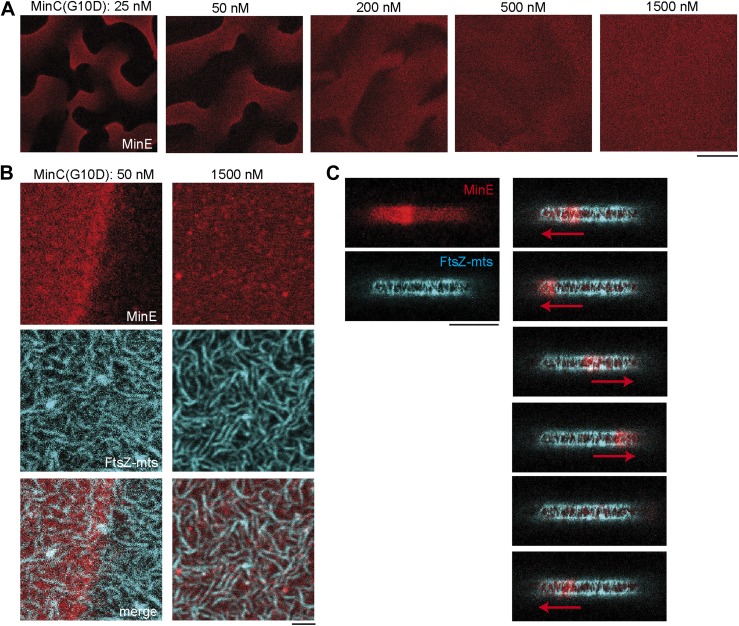
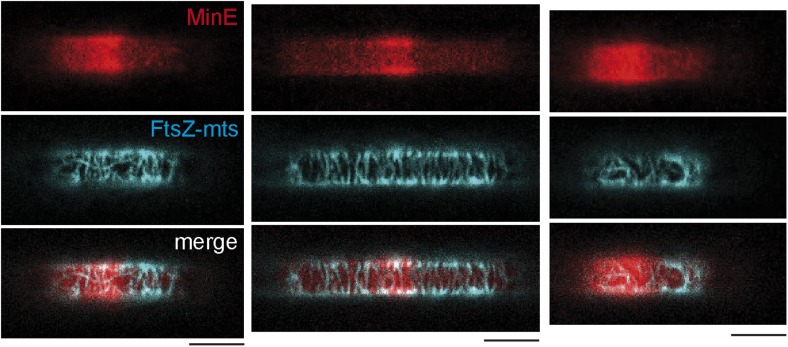
Figure 3. MinC-G10D does not inhibit polymerization of FtsZ-mts.
(A–C) FtsZ-mts (blue), MinD, MinE (5% labeled with Atto655: red) were reconstituted with the MinC-G10D mutant. Concentrations of MinD, MinE and FtsZ-mts are constant for all images. (A) Confocal images of Min protein pattern (MinE.Atto655: red) on flat supported membranes at different concentrations of MinC-G10D. High concentrations of MinC-G10D disturb Min protein patterns. The contrast between images is not comparable and is increased for higher concentrations of MinC-G10D, because the MinE intensity at the membrane decreased with higher concentrations of MinC-G10D. Scale bar: 50 µm. (B) Confocal images of the Min system (MinE.Atto655:red) and FtsZ-mts (blue) on flat membranes at different concentrations of MinC-G10D demonstrate that MinC-G10D is inefficient in disturbing FtsZ-mts networks. Scale bar: 2 µm. (C, Figure 3—figure supplement 1) In cell-shaped micro compartments with MinD, MinE, FtsZ-mts and 50 nM MinC-G10D the Min system (MinE.Atto655: red) oscillates from pole-to-pole. However MinC-G10D does not inhibit polymerization of FtsZ-mts (blue) at the compartment poles. Time between frames: 90 s, scale bar: 10 µm.