Figure 1. Diversity of Riboswitches and Mechanisms of Gene Control in Bacteria.
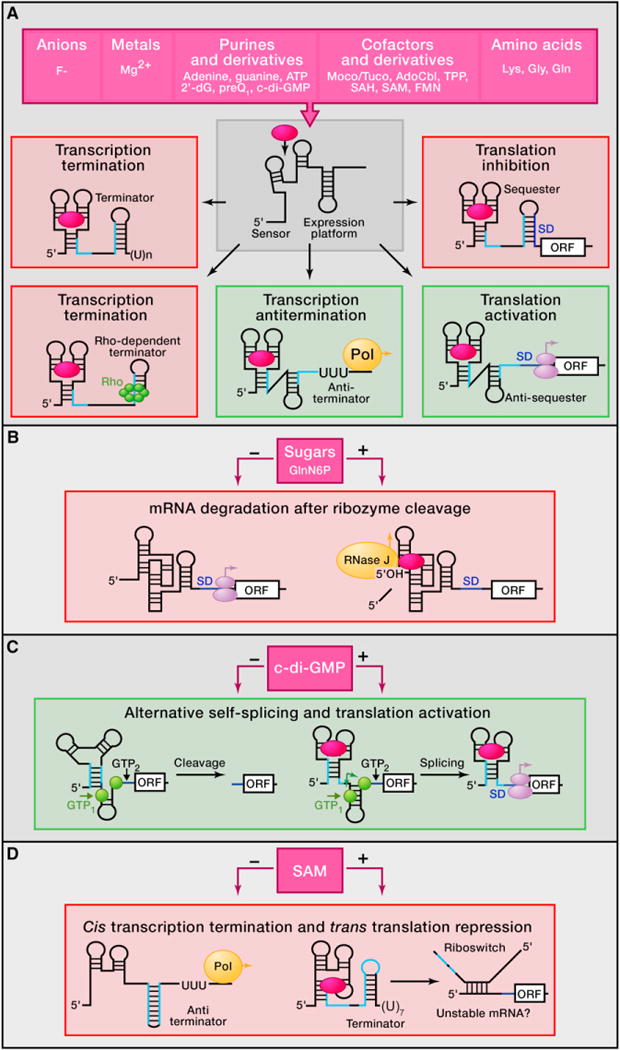
Mechanisms of modulation of gene expression are highly divergent in prokaryotes and involve control of transcription, translation, splicing, and mRNA stability.
(A) Various metabolites (violet, top) present in cells above threshold concentration can be directly sensed and specifically bound by sensor domains of riboswitches (white). Entrapment of a metabolite stabilizes the ligand-bound conformation of the riboswitch sensor and typically induces structural changes in the adjacent region (expression platform) that bears structural elements that stimulate (green) or repress (pink) gene expression. GlnN6P, glucosamine-6-phosphate; 2′-dG, 2′ -deoxyguanosine; preQ1, pre-queuosine-1; c-di-GMP, cyclic di-guanosyl-5′-monophosphate; Moco/Tuco, molybdenum and tungsten cofactors; SAH, S-adenosyl-L-homocysteine. Left: metabolite binding most often prevents formation of the antiterminator hairpin (complementary RNA regions in light blue) and promotes formation of the alternative Rho-independent termination hairpin (middle) or Rho binding site (bottom) that causes premature transcription termination. Center: in some cases, bound metabolites stabilize the antiterminator hairpin that allows RNA polymerase (Pol) to complete transcription of the gene (bottom). Right: expression of open reading frames (ORF) can be repressed by sequestration of the ribosome entry site (RBS or Shine-Dalgarno sequence, SD, dark blue) and blockage of translation initiation (middle). Metabolite binding to some riboswitches facilitates formation of the SD antisequester hairpin that opens up SD for ribosome binding and translation initiation (bottom).
(B) In gram-positive bacteria, bound GlcN6P induces cleavage by the glmS riboswitch-ribozyme in the 5′ UTR. The 5′-OH of the resulting fragment stimulates degradation of the message by RNase J.
(C) C. difficile exploits an allosteric ribozyme-riboswitch that combines self-splicing and translation activation. Left: in the absence of c-di-GMP, the intron uses GTP cleavage site 2 (black triangle, GTP2), thus yielding RNAs with truncated SDs that are not expressed. Right: binding of c-di-GMP to the riboswitch in the presence of GTP promotes cleavage at site 1 (green triangle, GTP1) and self-excision (green arrows) of the group I self-splicing intron using splicing sites shown in green circles. This self-cleavage brings together two halves of the SD, and the resulting mRNA is efficiently translated.
(D) In L. monocytogenes, the SreA and SreB riboswitches form antiterminator hairpins and allow normal transcription in the absence of SAM. Binding of SAM causes transcription termination. The resulting mRNA fragments base pair with the 5′ UTR of the mRNA and functions in trans as an antisense sRNA that destabilizes the target transcript, thus reducing protein production.
