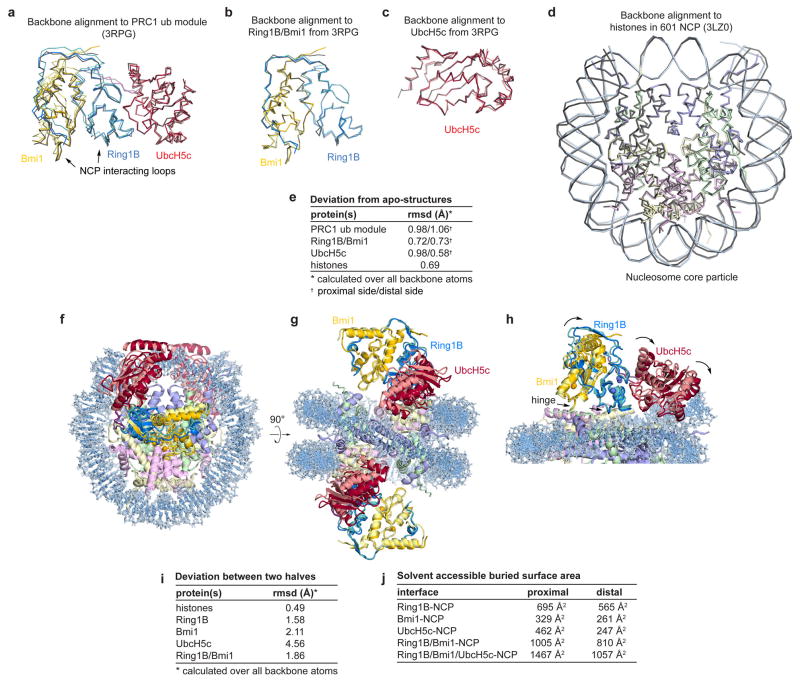
Extended Data Figure 1. Alignment of PRC1 ubiquitylation module and nucleosome core particle with previously determined sub-structures and comparison of the two halves of the structure.
a, Alignment of PRC1 ubiquitylation model from the proximal (darker colors) and distal (lighter colors) halves of the PRC1 ubiquitylation module-nucleosome core particle structure with the previously determined structure of the PRC1 ubiquitylation module alone22 (grey, PDB ID 3RPG) using all backbone atoms. Similar alignment using b, Ring1B/Bmi1 subcomplex or c, UbcH5c only. d, Alignment of the nucleosome core particle from the PRC1 ubiquitylation module-nucleosome core particle complex (colored) and the previously determined nucleosome core particle complex25 (PDB ID 3LZ0) containing the identical 601 nucleosome positioning sequence. e, Root mean square deviation (rmsd) for each of the alignments calculated over all backbone atoms. f–g, Orthogonal views of an alignment of two copies of the complex following rotation of one copy about the pseudo-two-fold axis of symmetry of the nucleosome. This was accomplished by simultaneously aligning each histone in one copy to its symmetry related counterpart in the other copy. For visualization, the PRC1 ubiquitylation module from one copy of the structure is depicted with lighter colors. h, Zoomed view of the alignment showing PRC1 ubiquitylation module-nucleosome core particle interface. In panel h the proximal and distal PRC1 ubiquitylation modules are shown in dark and light colors, respectively. Hinge indicated by arrows. i, rmsd for each of the components following alignment of the two halves as described above. j, Buried surface area calculations for the indicated interfaces on the proximal and distal sides of the structure.