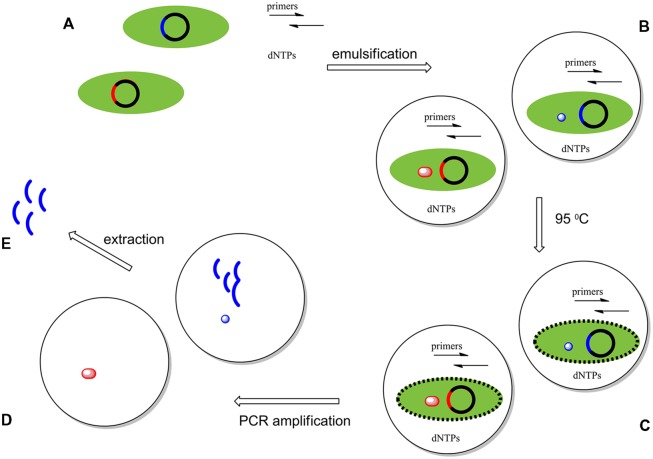
FIGURE 3.
Compartmentalized self replication (CSR) system experiments start with the creation of a library of genes encoding variants of a polymerase. Members of this library are introduced into E. coli cells by electroporation. Here, just two variant genes (red and blue) are represented. These genes drive the expression of mutant polymerases in each E. coli cell, each of which is isolated in its own water-in-oil-emulsion droplet. (B) The first cycle of PCR breaks the cell wall of the E. coli, exposing the expressed polymerase molecules and their gene to the contents of a water droplet containing all of the necessary components necessary for a PCR amplification: (i) primers, (ii) dNTPs, (iii) a mutated gene of the polymerase, and (iv) the enzyme expressed by this gene (C). During PCR, any polymerases active under the selective pressure (blue) amplify their respective genes, enriching the pool of mutants having the desired properties; inactive polymerases (red) fail to do so (D). The emulsion is then broken and the amplified genes enriched in those encoding polymerases having the desired behaviors are extracted and inserted in a plasmid vector [circular DNA; E]. These then enter the cycle of selection again (A). After repeating these cycles an enriched pool of variants of the original gene are produced.