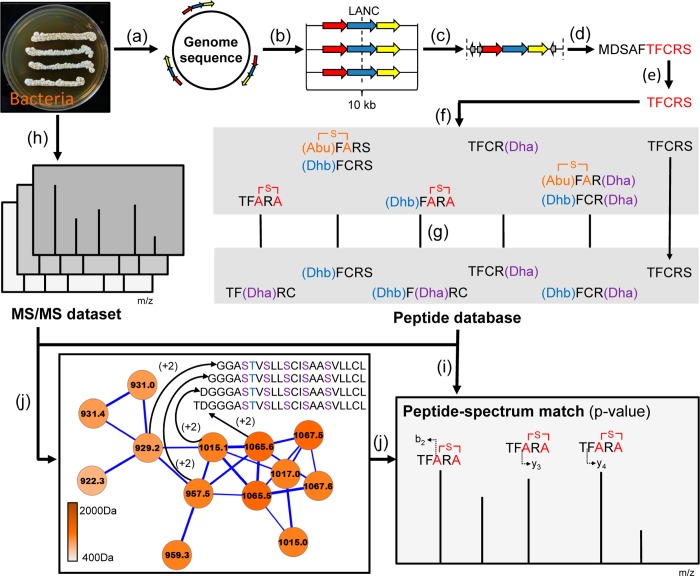
Figure 1.
Workflow implemented in the RiPPquest algorithm for automated peptidogenomics of RiPPs. (a) Prediction of lanthipeptide gene clusters in microbial genome sequence. (b) Generation of 10 kb windows centered at LANC-domain of gene clusters. (c) Prediction of ORFs in each gene cluster. (d) Selection of all candidate precursor peptides ORFs <100 aa. (e) Generation of candidate core peptides via C-terminal half of each selected ORF. (f) Generation of all biosynthetic and gas phase products of each core peptide, exemplified by peptide TFCRS. (g) Generation of MS/MS peptide database of predicted lanthipeptide products. (h) MS/MS analysis of microbial extract. (i) Matching of MS/MS data with MS/MS lanthipeptide spectral database with computed p-values. (j) Molecular network analysis of MS/MS data to identify peptide homologues and to confirm PSMs.