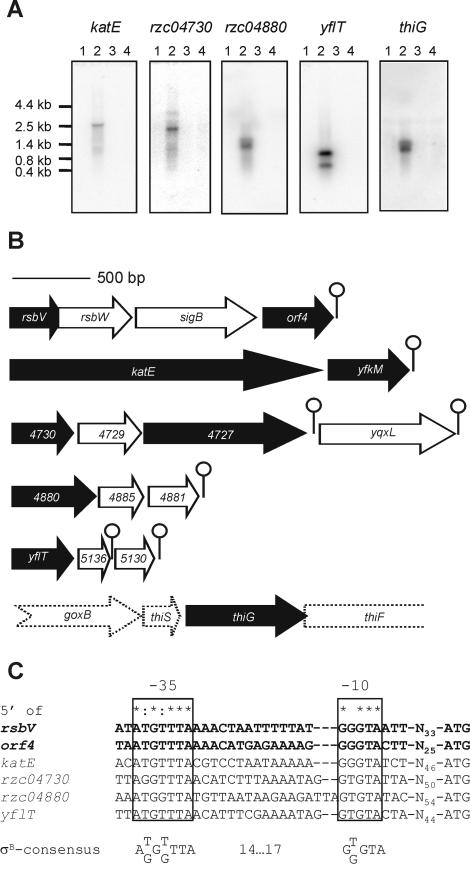
FIG. 3.
Transcriptional analysis of σB-dependent genes in B. cereus. (A) Northern blot analysis of transcription of σB-dependent genes in B. cereus. Total RNAs were extracted from B. cereus ATCC 14579 and B. cereus FM1400 cells during mid-exponential growth in BHI medium (lanes 1 and 3, respectively) and after a 10-min exposure to 42°C (lanes 2 and 4, respectively). 32P-labeled internal PCR products of the different genes were used as probes. Northern blot analyses with probes corresponding to yfkM and rzc04727 gave identical results as the Northern blot analyses with probes corresponding to katE and rzc04730, respectively (data not shown). Marker sizes (in kilobases) are indicated. (B) Operon structure of σB-dependent genes in B. cereus. The arrows represent open reading frames and indicate their orientations and sizes. Black arrows correspond to genes that were identified on the basis of 2D-E experiments. White arrows denote genes that are cotranscribed. Predicted stem-loop structures are indicated as lollipop structures. The three-letter codes of the genes, or when no such code exists, the B. cereus genome sequence code is indicated. The common part (RZC0) of the B. cereus genome sequence designations was omitted because of lack of space. The dashed arrows of the genes surrounding thiG indicate that it is not clear if these genes are cotranscribed with thiG. (C) Alignment of predicted σB-dependent promoter sequences. The −35 and −10 regions are indicated. The spacing to the start codon is also specified. The promoters 5′ of rsbV and orf4 are shown in bold to indicate that these have been experimentally defined (52). Completely conserved residues are indicated with asterisks. Residues that are conserved in five of the six promoter sites are indicated with colons. The extracted σB promoter consensus sequence is also indicated.