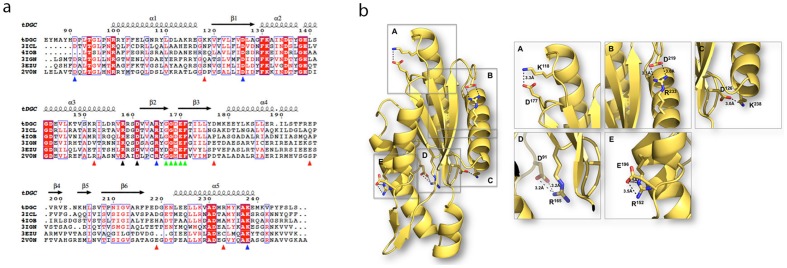
Figure 5. Structure-based sequence alignment of selected DGCs and salt-bridges unique to tDGC.
(a) Sequence alignment between tDGC from Thermotoga maritima (this work), mesophilic DGC domains (3ICL, 4IOB, 3IGN and 3EZU). The sequence of the structurally characterized PleD (2V0N) from Caulobacter crescentus is also included for comparison. The secondary structure elements of tDGC are displayed above the sequences. Strictly conserved residues are in red boxes and chemically similar residues are colored in red. Residues D126 and D169 of tDGC are involved in divalent metal coordination. The A- and I-site residues are highlighted with green and black triangles respectively. The blue triangles indicate salt-bridge forming residues that are conserved across the five proteins. Red triangles indicate residues unique to tDGC involved in salt bridge formation. (b) Mapping of the salt bridges unique to the thermostable enzyme onto the 3D structure of tDGC: Lys118-Asp177, Arg233-Asp219 and Arg152–Glu196 were disrupted in the site-directed mutagenesis study.