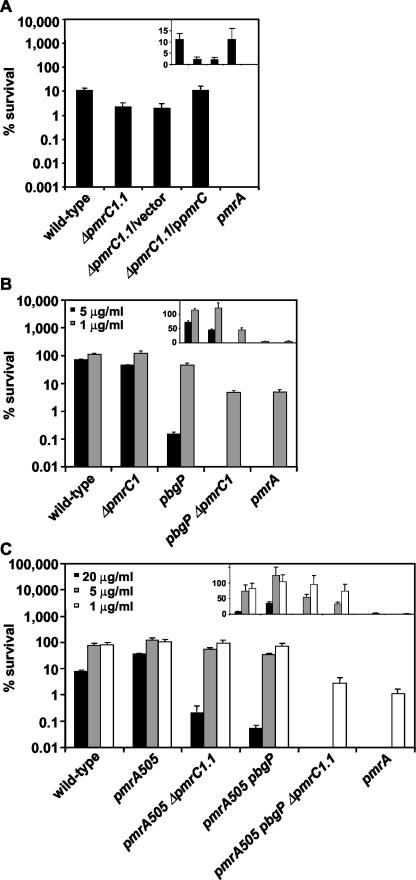
FIG. 3.
(A) Polymyxin B killing assay of wild-type (14028s), ΔpmrC1.1 (EG14590), ΔpmrC1.1/vector (EG14656), ΔpmrC1.1/ppmrC (EG14595), and pmrA (EG7139) strains grown to logarithmic phase in N-minimal medium, pH 5.8, with 10 μM MgCl2. Polymyxin B was added to a final concentration of 10 μg/ml, and the bacteria were incubated for 1 h at 37°C. The samples were diluted in PBS and plated on LB agar plates to determine the numbers of CFU. Survival values given are relative to those of PBS-treated samples. The ΔpmrC1.1 (EG14590) and ΔpmrC1.1/vector (EG14656) strains were significantly more sensitive to polymyxin B than was the wild-type (14028s) strain (P < 0.01). The complemented strain ΔpmrC1.1/ppmrC (EG14595) was significantly more resistant to polymyxin B than were strains ΔpmrC1.1 (EG14590) and ΔpmrC1.1/vector (EG14656) (P < 0.01). (B) Polymyxin B killing assay of wild-type (14028s), ΔpmrC1 (EG13927), pbgP (EG9241), pbgP ΔpmrC1 (EG14372), and pmrA (EG7139) strains grown and tested as described above, except that polymyxin B was added at final concentrations of 1 and 5 μg/ml. The difference in the polymyxin B (1 μg/ml) susceptibilities of strains pbgP ΔpmrC1 (EG14372) and pmrA (EG7139) was not statistically significant (P = 0.7), indicating that the pbgP and pmrC loci mediate PmrA-controlled polymyxin B resistance. (C) Polymyxin B killing assay of wild-type (14028s), pmrA505 (EG9492), pmrA505 ΔpmrC1.1 (EG14368), pmrA505 pbgP (EG9868), pmrA505 pbgP ΔpmrC1.1 (EG14369), and pmrA (EG7139) strains grown and tested as described for panel A, except that polymyxin B was added at 1, 5, and 20 μg/ml. Note the logarithmic scale (a linear scale is used in the insets) on the y axis. The data correspond to mean values from three independent sets of experiments performed in duplicate. The data demonstrate that the inactivation of the pmrC gene increases the susceptibility of cells to polymyxin B and that a pbgP ΔpmrC1 double mutant exhibits the same level of polymyxin B susceptibility as the pmrA null mutant.