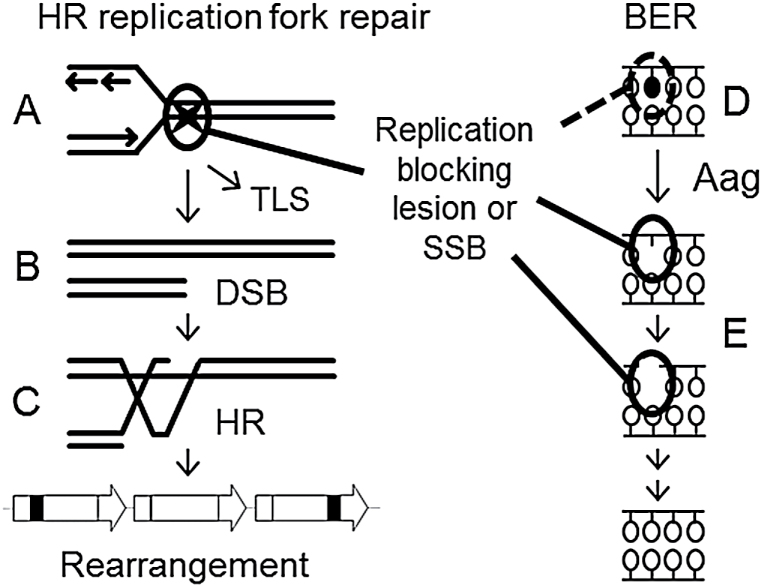
Fig. 6.
Model for the interaction of DNA damage, cell proliferation and Aag-initiated BER on HR-driven sequence rearrangements. (A–C) A replication blocking DNA lesion or SSB (denoted by a star) can induce a sequence rearrangement through replication fork breakdown. (A) When encountered by the replication fork, the lesion may be bypassed by translesion synthesis (TLS) or (B) may lead to replication fork breakdown and the formation of a one-ended DSB. (C) The DSB is then repaired by HR, but a misalignment between repeated sequences can produce a sequence rearrangement. (D) Replication blocking DNA damage, such as the Aag substrate 3-methyladenine, synergizes with cell proliferation to induce replication fork breakdown and the formation of HR-driven rearrangements. (E) BER intermediates are formed after the Aag-mediated excision of both replication blocking and innocuous base lesions. BER intermediates can also block replication and induce rearrangement formation. Thus, an imbalance of Aag activity relative to the activities of downstream BER enzymes can modulate the formation of HR-driven rearrangements induced by DNA methylation damage. SSB, single strand break.