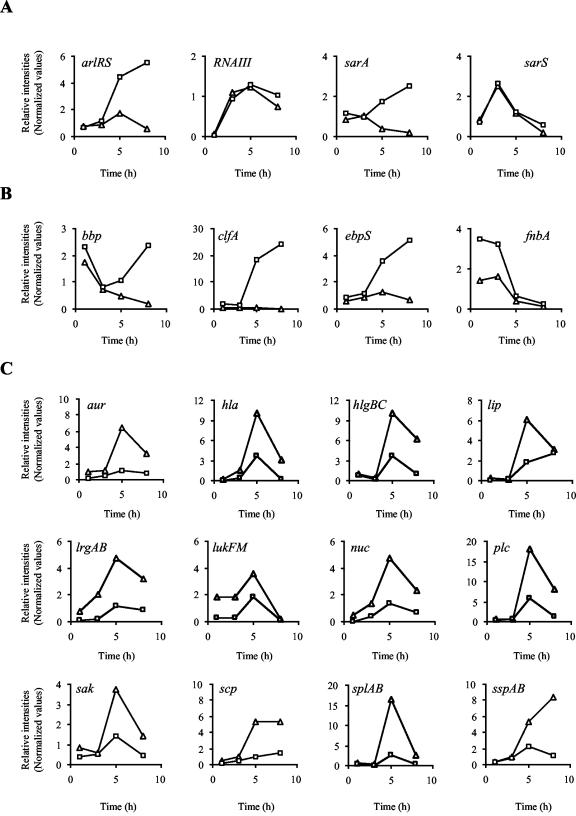
FIG. 4.
Transcription profiles of ORFs influenced by σB. Transcript levels for Newman (□) and IK184 (▵) cells sampled at different time points of growth (x axes). Data points were plotted as relative intensity values (y axes). (A) Influence of σB on known regulatory elements. arlR, autolysis-related locus regulator protein (response regulator); arlS, autolysis-related locus sensor protein (histidine kinase); RNAIII, effector molecule of the agr locus; sarA, staphylococcal accessory regulator A; sarS, staphylococcal accessory regulator S. (B) Adhesions factors upregulated by σB. bbp, bone sialoprotein-binding protein; clfA, clumping factor A; ebpS, elastin binding protein S; fnbA, fibronectin binding protein A. (C) Exoproteins and toxins downregulated by σB. aur, zinc metalloprotease aureolysin; hla, α-hemolysin; hlgBC, γ-hemolysin components B and C; lip, lipase; lrgAB, holin-like proteins LrgA and LrgB; lukF, synergohymenotropic toxin precursor; lukM, leukocidin chain precursor; nuc, staphylococcal nuclease; plc, 1-phosphatidylinositol phosphodiesterase precursor; sak, staphylokinase precursor (protease III); scp, staphopain; splAB, serine proteases SplA and SplB; sspA, staphylococcal serine protease (V8 protease); sspB, cysteine protease SspB.