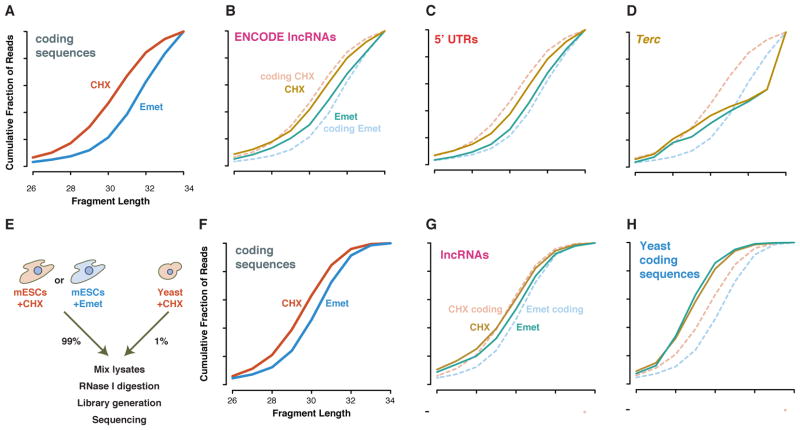
Figure 2. Elongation inhibitors shift ribosome footprint sizes.
(A) Cumulative length distribution shows ~1 nt larger footprints on annotated coding sequences from emetine-versus cycloheximide-treated cells (Ingolia et al., 2011). (B) LncRNA and (C) 5′ UTR footprints from transcripts passing the FLOSS cutoff show a similar length shift, whereas background from (D) classical non-coding RNAs do not. (E) Experimental design with e cycloheximide-treated yeast polysomes as an internal standard for nuclease digestion and library generation. (F) Annotated coding seuqences and (G) lncRNAs again show larger footprints in emetine-treated cells. (H) Cycloheximide-stabilizied footprints are not larger in the emetine-treated mESC lysate sample.