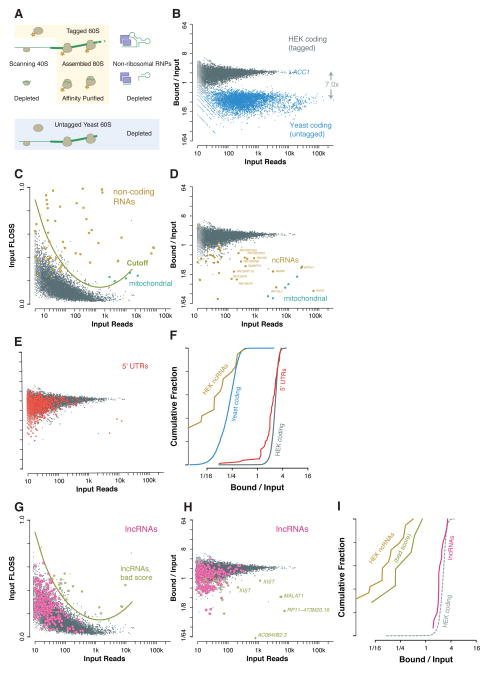
Figure 3. Ribosome affinity purification separates 80S footprints from background RNA.
(A) Schematic showing that affinity purification of tagged 60S ribosome subunits recovers 80S footprints but depletes background from non-ribosomal RNPs, potential scanning 40S footprints, and footprints of untagged yeast 80S ribosomes are also depleted. (B) Human ribosome footprints are retained during ribosome affinity purification while yeast ribosome footprints (excepting the yeast biotin carrier ACC1) are depleted. (C) Fragment length analysis of nuclear and mitochondrial coding sequences and of functional non-coding RNAs in HEK cells. A fragment length score cutoff based on extreme outliers relative to coding sequences excludes background fragments. (D) Ribosome footprints are retained during ribosome affinity purification while mitochondrial footprints and non-coding RNAs are depleted. (E, F) Ribosome footprints on 5′ UTRs are retained during affinity purification of the 60S ribosomal subunit. (G) Fragment length analysis of ENCODE lncRNAs, identifying a small number of transcripts with likely non-ribosomal contamination. (H, I) Ribosome footprints on lncRNAs are retained during ribosome affinity purification, whereas many sources of non-ribosomal contamination, including the nuclear non-coding RNA XIST, are depleted.