Figure 3.
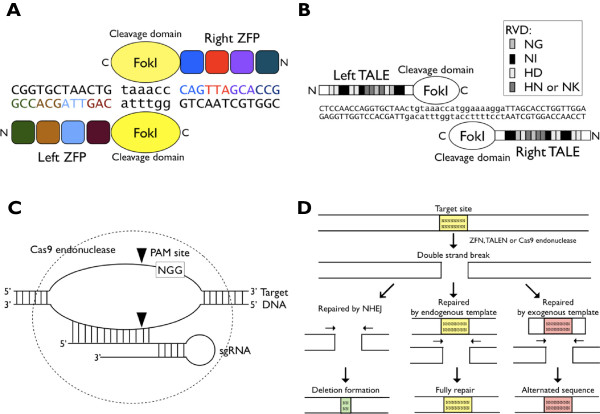
Site-specific endonucleases. Three classic site-specific endonucleases including ZFN, TALEN and CRISPR/Cas9 are shown. (A) ZFN simply consists of a Zinc-finger protein (ZFP) fused to Fok1 endonuclease. The sequence composition of the α-helix in the zinc-finger determines the nucleotide binding specificity of the ZFP. As a result, a ZFP chain can be created by joining a few ZFPs together and generating high specificity, allowing Fok1 endonuclease to accurately cleave DNA at the target site. (B) The C-terminal end of a TALE contains a Fok1 endonuclease for DNA cleavage. The central part of the TALE contains a number of almost similar repeats that mediate specific binding to target loci in the genome, and each of these repeats specifically binds to one base of the target DNA via two amino acids named repeat variable di-residues (RVDs), including NG, NI, HD and HN (or NK) for recognizing one of the four different nucleotides: T, A, C and G, respectively [53, 54]. (C) Cas9 forms a sequence-specific endonuclease when complexed with the sgRNA. The Cas9/sgRNA complex then recognizes the targeted sequence, 20-bp in length, ending with two guanines (NGG) called the PAM site. Cleavage occurs on both strands upstream of the PAM sites. (D) The DSB is first induced by ZFN, TALEN or Cas9 endonuclease and then repaired by three possible mechanisms. When repaired by NHEJ, random deletions would occur at the site (left). When the repair is done by the endogenous template within the genome, the sequence would be fully repaired (middle). If an exogenous modified template is added, the sequence could be altered after repair, which is regarded as the gene editing (right).
