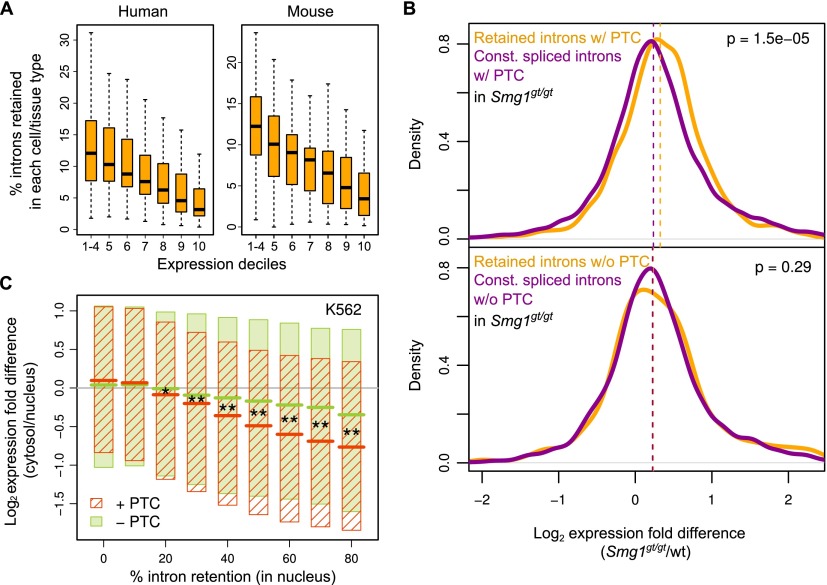
Figure 4.
Global regulation of mRNA levels through IR. (A) Box plots showing distributions of percentages of total human and mouse introns detected as retained (PIR ≥ 10) in transcripts sorted into 10 different expression level bins (deciles, with deciles 1–4 averaged). (B) Distributions of expression difference between Smg1gt/gt and wild-type MEFs for transcripts harboring introns predicted to introduce a PTC that can trigger NMD upon retention (top), and for transcripts harboring introns that are predicted not to introduce a PTC (bottom). In each panel, retained (PIR ≥ 10) and constitutively (PIR < 2) spliced introns are compared. P-value indicates significance of expression change difference (one-sided Mann-Whitney U test). (C) Expression difference between cytosolic and nuclear fractions of K562 cells for transcripts that do or do not contain PTC-introducing introns, as measured at different PIR thresholds in the nuclear fraction (see also Supplemental Fig. S6C). Shaded boxes indicate upper and lower quartiles of distributions of expression level differences, and colored lines indicate median values. Expression-level differences for transcripts harboring retained introns that do or do not introduce PTCs are indicated by red and green, respectively. Asterisks indicate significance of difference between median values for expression level differences for transcripts with and without PTC-introducing retained introns (*) P < 0.05; (**) P < 0.001 (one-sided Mann-Whitney U tests after Bonferroni correction). See also Supplemental Figure S7B.