Abstract
The endemicity of highly pathogenic avian influenza (HPAI) A(H5N1) viruses in Asia has led to the generation of reassortant H5 strains with novel gene constellations. A newly emerged HPAI A(H5N8) virus caused poultry outbreaks in the Republic of Korea in 2014. Because newly emerging high-pathogenicity H5 viruses continue to pose public health risks, it is imperative that their pathobiological properties be examined. Here, we characterized A/mallard duck/Korea/W452/2014 (MDk/W452(H5N8)), a representative virus, and evaluated its pathogenic and pandemic potential in various animal models. We found that MDk/W452(H5N8), which originated from the reassortment of wild bird viruses harbored by migratory waterfowl in eastern China, replicated systemically and was lethal in chickens, but appeared to be attenuated, albeit efficiently transmitted, in ducks. Despite predominant attachment to avian-like virus receptors, MDk/W452(H5N8) also exhibited detectable human virus-like receptor binding and replicated in human respiratory tract tissues. In mice, MDk/W452(H5N8) was moderately pathogenic and had limited tissue tropism relative to previous HPAI A(H5N1) viruses. It also induced moderate nasal wash titers in inoculated ferrets; additionally, it was recovered in extrapulmonary tissues and one of three direct-contact ferrets seroconverted without shedding. Moreover, domesticated cats appeared to be more susceptible than dogs to virus infection. With their potential to become established in ducks, continued circulation of A(H5N8) viruses could alter the genetic evolution of pre-existing avian poultry strains. Overall, detailed virological investigation remains a necessity given the capacity of H5 viruses to evolve to cause human illness with few changes in the viral genome.
Keywords: avian influenza virus, genetic evolution, HPAI A(H5N8), migratory waterfowl, reassortment
INTRODUCTION
Since the mid-1990s, highly pathogenic avian influenza (HPAI) A(H5N1) viruses have ravaged domestic poultry in Asia. These viruses' unprecedented spread towards Africa and Europe since 2003 gave rise to at least 10 distinct phylogenetic clades based on the phylogenetic position of their hemagglutinin (HA) genes.1 During the same period, the World Health Organization recorded more than 640 cases of human infections worldwide that were due to direct contact (DC) or repeated extensive exposure to infected birds and reported an approximately 60% mortality rate among those infected.2 The endemicity of HPAI A(H5N1) in poultry and wild birds also led to the generation of novel reassortant H5 strains that inherited neuraminidases (NAs) and internal gene constellations from other prevailing subtypes of avian viruses circulating in the field.3,4,5,6,7,8,9
In the Republic of Korea (Korea), at least four outbreaks due to HPAI A(H5N1) viruses of different clade groups have occurred.10,11,12,13 In each of these instances, wild waterfowl played a significant role in the emergence and spread of the respective viral pathogens. Because immediate and effective response measures were taken, none of these events managed to establish the causative virus in domestic poultry. In mid-January 2014, HPAI H5 viruses emerged anew in Korean domestic poultry, causing outbreaks in breeding ducks and chickens.14 Domestic outbreaks were also associated with die-offs of wild birds near the first reported poultry cases in Jeonbuk province, leading to speculation that migratory waterfowl were the source of infection. The causative virus was later identified as a novel HPAI A(H5N8) reassortant,15 eventually resulting in the culling of more than 12 million poultry birds. Because the public health risk of the novel HPAI A(H5N8) virus to animals and humans remains uncertain, we aimed in the present study to evaluate the pathogenic potential and pathobiological features of the virus in various animal models.
MATERIALS AND METHODS
Ethics statement
All animal experiment protocols performed in this study followed general animal care guidelines mandated under the Guidelines for Animal Use and Care of the Korea Center for Disease Control (K-CDC). They were approved by the Medical Research Institute (approval NO CBNU-IRB-2012-GM01) and by the Laboratory Animal Research Center (approval NO CBNUA-074-0904-01), which is a member of the Institutional Animal Care and Use Committee of Chungbuk National University, and were performed in partnership with Bioleaders Corp. (permit NO BLS-ABSL-10-003). Ex vivo experiments involving human respiratory tissues were conducted using protocols approved by the Ethics Committee of the Faculty of Medicine at Chungbuk National University in Cheongju, Korea. The experimental protocol was comprehensively explained to patients undergoing tissue biopsies and duly signed written consent forms were obtained.
Cells
Madin–Darby canine kidney (MDCK) cells (American Type Culture Collection, Manassas, VA, USA) were grown and maintained in Eagle's minimum essential medium with Earle's salts (Lonza, Basel, Switzerland) containing 5% fetal bovine serum (Gibco Life Technologies, Grand Island, NY, USA). Primary normal human bronchial epithelial (NHBE) cells were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA) and differentiated as previously described.16 All cells were incubated at 37 °C in 5% CO2 until use. Primary chicken embryonic lung (CELu) and liver (CELi) cells were isolated and prepared from specific pathogen-free (SPF) 15-day-old White Leghorn chicken embryos (CAVac Lab. Co., Ltd., Daejeon, Korea) as described elsewhere17 using a protocol approved by the Medical Research Institute of Chungbuk National University (approval NO CBNU-IRB-2012-GM01). Upon isolation, tissues were minced, trypsinized and cultured at 37 °C in 5% CO2 in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 2 mM glutamine and antibiotics. Cell viability was assessed via Trypan blue exclusion assays and was not less than 90% for any preparation.
Viruses
Except for A/California/07/2009(H1N1) (CA/07(H1N1)), which was propagated in MDCK cells, the HPAI H5 viruses were isolated from wild bird fecal samples in the winter seasons of 2006–2007 [A/environment/Korea/W149/2006 (En/W149(H5N1))], 2010–2011 [A/mallard duck/Korea/W401/2011 (MDk/W401(H5N1))] and 2013–2014 [A/mallard duck/Korea/W452/2014 (MDk/W452(H5N8))] and grown in SPF 10-day-old embryonated chicken eggs. Supernatants (allantoic fluids and cell culture) were harvested at 48 h post-infection (hpi), aliquoted into cryovials (1 mL each), and stored at −80 °C until use. Stock viral titers were determined by 50% egg infectious dose (EID50) and 50% tissue culture infectious dose (TCID50) end-point titrations.18 All experiments with HPAI H5 and A(H1N1) viruses, including virus titrations in biological samples and receptor-binding assays, were conducted in an enhanced biosafety level 3 (BSL3+) containment facility as approved by K-CDC.
Genetic and phylogenetic analyses
Sequences were prepared and analyzed as previously described.19 Briefly, gene sequences of MDk/W452(H5N8) were obtained by Cosmo Genetech (Seoul, Korea) using an ABI 3730XL DNA sequencer (Applied Biosystems, Foster City, CA, USA). Sequences were analyzed and compiled with DNAStar 5.0 (DNAStar, Madison, WI, USA); closely related viruses were identified by basic local alignment search tool analysis. Phylogenetic trees were built by aligning published reference avian influenza virus sequences obtained from wild birds, domestic poultry and humans that are available in GenBank together with the closely related avian virus sequences obtained from the basic local alignment search tool results. Full genome sequences were aligned and bootstrapped in Clustal X,20 and phylogenetic trees were viewed using NJ Plot.21 The number of bootstrap replications was set to 1000, and major tree branches were labeled with bootstrap values (>60%) for reference. Branch lengths are proportional to sequence divergence and can be measured relative to the scale bar included in the phylogenetic trees (Supplementary Figure S1). MDk/W452(H5N8) was assigned GenBank accession numbers KJ746108-K746115.
Virus titrations
Virus titers in virus stocks, oropharyngeal and cloacal swabs, nasal washes, homogenized/clarified organ tissue samples and culture supernatants were determined by performing endpoint titrations in 10-day-old embryonated chicken eggs, monolayers of MDCK cells or both. Eggs or cells were inoculated with 10-fold serial dilutions of each sample in 1× phosphate-buffered saline solution or fetal bovine serum-free media containing antibiotics. After 48-h incubation at 35 °C, the presence of viruses was detected by a standard HA assay using 0.5% chicken erythrocytes. Mean virus titers were expressed as log10EID50 or as log10TCID50 per unit-sample (mL or g) tested. The limit of virus detection was set at <0.7 or 2.5 log10 per unit-sample tested.
NA inhibitors and NA-Star neuraminidase inhibitor resistance detection test
The NA inhibitors oseltamivir carboxylate and zanamivir were purchased from Toronto Research Chemicals, Inc. (Toronto, Ontario, Canada), and peramivir was acquired from Green Cross (Yongin, Kyunggi-do, Korea). The chemiluminescent NA inhibitor assay was conducted using the commercially available NA-Star kit (Applied Biosystems). NA-Star substrate was used at a final concentration of 100 µM. Briefly, 25 µL of half-log dilutions of neuraminidase inhibitors (0.03–1000 nM) in NA-Star Assay buffer were added to each well of a white 96-well microplate plate, 25 µL of virus dilution was added and plates were pre-incubated at 37 °C for 20 min. Diluted NA-Star (0.01 mM; 10 µL) was then added to each well, and the plate was incubated for 10 min at room temperature. Finally, 60 µL of NA-Star Accelerator was added to each well, and luminescence at 450 nm was read immediately (1.0 s/well). The 50% inhibitory concentration (IC50) values were determined by using GraphPad Prism™ software (v5) for non-linear regression analysis. For comparison and to serve as a positive control, the 2009 pandemic virus A/California/04/2009 was modified by introducing the Oseltamivir-resistance marker His274Tyr into its NA gene (CA/04(H1N1)H274Y) through site-directed mutagenesis and then rescued via reverse genetics.16
Growth kinetics of virus in vitro and plaque formation assay
MDCK, differentiated primary NHBE, and primary CELu and CELi cells were infected in triplicate in six-well plates with MDk/W452(H5N8), En/W149(H5N1), MDk/W401(H5N1) or CA/07(H1N1) at multiplicities of infection of 0.001, 0.1 or 0.01, respectively. After incubation at 35 °C for 45 min to 1 h, the viral inocula were replaced with serum-free medium appropriate for each cell line. Viral growth rates in all cells were determined three times in duplicate at 35 °C in the absence of L-1-tosylamido-2-phenylethyl chloromethylketone-treated trypsin. Cell culture supernatants were harvested at 6, 12, 24, 48 and 72 h hpi for virus titration in MDCK cells as specified above. Phenotypic characterization by plaque assay in the presence or absence of L-1-tosylamido-2-phenylethyl chloromethylketone-treated trypsin was carried out in six-well plates of MDCK cells infected with serial 10-fold dilutions of the viruses. Plaque formations were visualized at 48 hpi [En/W149(H5N1)] or 72 hpi [MDk/W452(H5N8), MDk/W401(H5N1)].
Infection of human respiratory tissues ex vivo
Fresh human nasal mucosal epithelium and lung tissue fragments were surgically obtained from patients undergoing removal of respiratory tissues for screening for possible carcinoma. Only normal non-malignant tissue fragments that were not needed for clinical diagnosis were used for infection as previously described.16 Briefly, excised tissues were cubed (5 mm×4 mm×2 mm), rinsed and incubated in Roswell Park Memorial Institute 1640 medium (Gibco) with L-glutamine and antibiotics for 2–3 h at 35 °C in 5% CO2. MDk/W452(H5N8), En/W149(H5N1), MDk/W401(H5N1) and CA/07(H1N1) (106 log10EID50/mL) were used to infect tissue sections (triplicates of 25 fragments per virus) in six-well plates. After 1 h of virus adsorption at 35 °C, the infected tissue sections were rinsed and segregated (five blocks each) in 12-well cell culture vessels containing 1 mL of Roswell Park Memorial Institute 1640 medium supplemented with 0.5 µg/mL L-1-tosylamido-2-phenylethyl chloromethylketone-treated trypsin solution (Sigma-Aldrich, St Louis, MO, USA), 1% bovine serum albumin and antibiotics. At 12, 24, 48 and 72 hpi, lung tissue sections were homogenized, and the homogenates were clarified by centrifugation. The titers of the viruses in tissue homogenates and cell culture supernatants were determined by EID50 assays. The remaining cubes (n=5 per virus) were fixed at 48 hpi and processed for immunohistochemical assays to detect Influenza A viral nucleoprotein (NP) (AbD Serotec, Kidlington, Oxford, UK) expression.
Receptor binding assays
Receptor affinity was determined in a solid-phase direct virus-binding assay as previously described.22 In brief, influenza viruses were bound to fetuin-coated plates at 4 °C overnight. Biotinylated glycans (α2,3′SL; α2,6′SL; or α2,6′SLN; Glycotech Corporation, Gaithersburg, MD, USA) were added to influenza-coated plates at varying dilutions and incubated for 4 h. Glycan binding was detected by adding horseradish peroxidase-conjugated streptavidin (Invitrogen, Carlsbad, CA, USA) followed by 3,3′,5,5′-tetramethylbenzidine substrate (Sigma-Aldrich, St Louis, MO, USA); the resulting absorbance at 450 nm was measured in a VICTOR3 1420 multilabel-counter plate reader (Perkin-Elmer, Waltham, MA, USA). The receptor binding specificities of HPAI H5 viruses were also determined in HA assays using 0.5% re-sialylated chicken red blood cells (cRBCs). For the HA assay, sialic acid residues were enzymatically removed from cRBCs by incubation of the cells with 50 mU Vibrio cholera neuraminidase (VCNA: Roche, San Francisco, CA, USA) at 37 °C for 1 h, the cells were then re-sialyated by incubation with either 2,6-(N)-sialyltransferase or 2,3-(N)-sialyltransferase (Sigma-Aldrich, St Louis, MO, USA) at 37 °C for 4 h.23
Animal experiments
Mice, ferrets, chickens, ducks, dogs and cats determined to be influenza-free by hemagglutination inhibition (HI) and NP-based enzyme-linked immunosorbent assay for currently circulating human and/or avian influenza viruses were used in our inoculation experiments; SPF chickens were purchased. The sample numbers were based on previous studies showing their sufficiency to identify a significant difference among virus groups.24,25,26,27,28 For the dogs and cats, two or three animals per group were used to evaluate tissue distribution and transmission, respectively. The experimental animals were arbitrarily allocated to groups without using a randomized selection method, and investigators were aware of the allocations. All experiments with HPAI A(H5N8) and A(H5N1) viruses were conducted in a BSL3+ facility. Enhanced BSL3 practices were used for the animal work, and approved institutional guidelines were followed. The facility is secured by procedures recognized as appropriate by the institutional biosafety officers and facility management of Bioleaders Corp. and by K-CDC government inspectors. Our research protocol and all animal experiments followed the Guidelines for Animal Use and Care of K-CDC and was approved by the Medical Research Institute of Chungbuk National University (approval NO CBNU-IRB-2012-GM01).
Experimental infection of chickens
Five-week-old female SPF white Leghorn chickens (CAVac Lab. Co., Ltd., Daejeon, Korea) were used in this study. To determine the LD50s (expressed as log10 EID50) of the viruses in these animals, groups of five chickens were oronasally inoculated with 10-fold serial dilutions containing 100–108 EID50/mL of MDk/W452(H5N8), En/W149(H5N1) or MDk/W401(H5N1). Pathogenicity indices were determined by intravenously inoculating groups of 10 chickens with 0.1 mL of a 1∶10 dilution of allantoic fluid containing virus. Groups of nine chickens were separately inoculated oronasally with 106 EID50/mL of virus. Three chickens per group were euthanized at 3 and 5 days post-infection (dpi) for virological examination. To test for virus transmission, six contact birds were cohoused with the remaining three infected hosts after 1 dpi and observed daily for 14 days. Tracheal and cloacal swabs were collected from the inoculated animals on alternate days and from contact birds every day. Virus titration assays of various organs and swab samples were done by EID50 assay.
Experimental infection of ducks
Groups of five 3-week-old domestic ducks (Anas platyrhynchos domesticus) were used to determine LD50s by inoculation with 10-fold serial dilutions containing 100–106 EID50/mL of MDk/W452(H5N8), En/W149(H5N1) or MDk/W401(H5N1). Twelve ducks were separately inoculated oronasally with 106 EID50/mL of virus. Three ducks per group were euthanized at 3 and 5 dpi for virological examination. To test for virus transmission, six contact birds were cohoused with the remaining six infected hosts after 1 dpi and observed daily for 14 days. Tracheal and cloacal swabs were collected from the inoculated birds every other day and from contact birds every day. Virus titrations of various organs and swab samples were determined by EID50 assay.
Experimental infection of mice
Baseline body weights of 6-week-old female BALB/c mice (Samtako, Seoul, Korea) were measured prior to infection. Groups of 10 mice per virus were intranasally inoculated with 102–107 EID50/50 µL of MDk/W452(H5N8), En/W149(H5N1) or MDk/W401(H5N1). Body weights and survival were recorded daily for 14 days. For virological and pathological examinations, 15 mice per group were intranasally infected with 105 EID50/50 µL of virus, and three mice per group were euthanized at 1, 3, 5, 7 and 9 dpi to examine the growth kinetics of the virus in mouse lung. Viral growth in extrapulmonary tissues was also determined at 3 and at 5 dpi. Virus titration in various organs was determined by EID50 assays.
Experimental infection of ferrets
Under anesthesia, groups of nine 16- to 18-week-old ferrets (Wuxi Sangosho Pet Park Co., Wuxi, China) were intranasally inoculated with 106 EID50/mL of MDk/W452(H5N8), En/W149(H5N1) or MDk/W401(H5N1). Three ferrets per group were euthanized at 3 and at 5 dpi for virological and pathological examination. At 1 dpi, the other three inoculated animals were individually paired by cohousing with a DC ferret; a respiratory-droplet (RD) contact animal was also housed in a wire frame cage adjacent to the infected ferret. The infected and RD contact ferrets were 5 cm apart, preventing direct and indirect contact between animals but allowing the spread of the influenza virus via aerosol droplets. The body weight and temperature of the animals were monitored every other day. Nasal washes were collected from the infected ferrets every other day for 9 days beginning at 1 dpi and daily from 1 day post-exposure from the contact ferrets. Virus titrations of nasal washes and various tissue organs were determined by EID50 assays.
Experimental infection of dogs and cats
Eight-week-old beagle dogs and domestic cats were used in this study (CAVac Lab. Co., Ltd., Daejeon, Korea). Under anesthesia, seven beagles and seven cats were intranasally inoculated with 106 EID50/mL of MDk/W452(H5N8). At 3 and 5 dpi, two animals per group were euthanized for virological and pathological examination. The three remaining animals in each group were individually paired with a naive DC host to determine transmission. Body weight and temperature were monitored every other day. Nasal swabs were collected from the infected animals every other day from 1 dpi and every day from 1 day post-exposure from the contact animals (for 7 days). Virus titrations of nasal swabs and various tissue organs were determined by EID50 assays.
Pathological examination
Excised tissues and animal organs were preserved in 10% phosphate-buffered formalin and processed for paraffin embedding. Sections from each organ and tissue underwent standard hematoxylin and eosin staining. Immunohistological staining with anti-Influenza A viral NP antigen (AbD Serotec, Kidlington, Oxford, UK) was also performed. Specific antigen–antibody reactions were visualized by 3,3′-diaminobenzidine tetrahydrochloride staining using the Vector Laboratories, Inc. system (Burlingame, CA, USA).
Cytokine and chemokine measurement
Broncheoalveolar lavage (BAL) fluid from influenza virus-infected mice was processed using a Luminex-based multiplex immunoassay kit (Procarta cytokine assay kit; Affymetrix, Santa Clara, CA, USA). Array analysis was performed using the Bio-Plex 200 system (Bio-Rad Laboratories, Hercules, CA, USA).
Statistical analysis
The differences among log10-transformed viral titers of different viruses were compared using Student's t-test or one-way analysis of variance. The statistical analyses were carried out using GraphPad Prism™ software (v5) (La Jolla, CA, USA).
RESULTS
Genetic characterization of novel A(H5N8) virus
We isolated four viruses from fecal samples of wild birds collected in Chungcheong provinces in February 2014. Alignment of the sequences reveals that they are 99.8%–100% identical to one another. A/mallard duck/Korea/W452/2014(H5N8), a representative virus hereinafter referred to as MDk/W452(H5N8), was isolated along the Mihocheon River, Chungcheongbuk-do in the mid-region of Korea. Sequence and phylogenetic analyses demonstrated that the HA, NA, basic polymerase 2 (PB2), and NP segments of MDk/W452(H5N8) were derived from HPAI A/duck/Jiangsu/k1203/2010(H5N8)-like virus, a proposed clade 2.3.4.6 H5 virus by virtue of its HA;8,29 the remaining basic polymerase 1 (PB1), acidic polymerase (PA), matrix (M) and non-structural (NS) segments were those of an A/duck/Jiangsu/1-15/2011(H4N2)-like virus30 (Supplementary Figure S1 and Supplementary Table S1). The NA gene was highly divergent (76.9% nucleotide homology) from the A(H10N8) avian virus (Supplementary Figure S1B), which has caused human infections in China.31 Moreover, all segments of the 2014 HPAI A(H5N8) virus were derived from strains that are resident in Eurasian migratory wild birds, and there were no indications of the A(H9N2)-like segments that were found in human-infecting A(H7N9) and A(H10N8) avian viruses in China.31,32
Lee et al.15 recently reported the detection of at least two genotypically distinct A(H5N8) viruses [A/breeder duck/Korea/Gochang1/2014, BDk/Gochang1(H5N8) and A/Baikal Teal/Korea/Donglim3/2014, BTl/Donglim3(H5N8)] these viruses were present in wild and domestic birds at the site of the initial outbreaks. Comparison of the full genome sequence of MDk/W452(H5N8) with those of BDk/Gochang1(H5N8) and BTl/Donglim3(H5N8) to understand their genetic relationships revealed that MDk/W452(H5N8) has a stronger genetic relationship (99.8%–100% sequence identity) to BTl/Donglim3(H5N8) (Supplementary Table S1), whose PB1, PA, M and NS genes originated from A/duck/Eastern China/1111/2011(H5N2)-like virus. The sequence identity of MDk/W452(H5N8) to BDk/Gochang1, whose PB2 and NS segments resemble those of an avian A(H11N9)-like virus, was only approximately 87%–97.7%. Notably, the parental A/duck/Jiangsu/1-15/2011(H4N2) (this study) and HPAI A/duck/Eastern China/1111/2011(H5N2) strains are >99% identical in all segments, differing only in HA.30
Although the H5 HA cleavage motif site of MDk/W452(H5N8) bears polybasic residues denoting a high-pathogenicity phenotype in chickens,15 molecular modifications that are known to promote infection of mammalian hosts are not present in this virus22,33,34,35 (Supplementary Table S2). An Asn66Ser substitution in the alternatively spliced PB1-F2 protein (PB1-F266S) that is known to contribute to virulence and facilitate secondary bacterial infections36,37 was noted, but no other mammalian-adaptive molecular determinants were observed in the viral genome.38,39 While the deduced non-structural 1 (NS1) protein harbored the Pro42Ser mutation associated with increased virulence in mice,40 seven additional amino-acid residues were also observed at the C-terminal region (Supplementary Table S2). MDk/W452(H5N8) may resist amantadine-derived compounds due to a matrix 2 ion-channel protein Ser31Asn mutation, but may have retained sensitivity to NA inhibitors.41,42
Biological properties of the HPAI (H5N8) virus
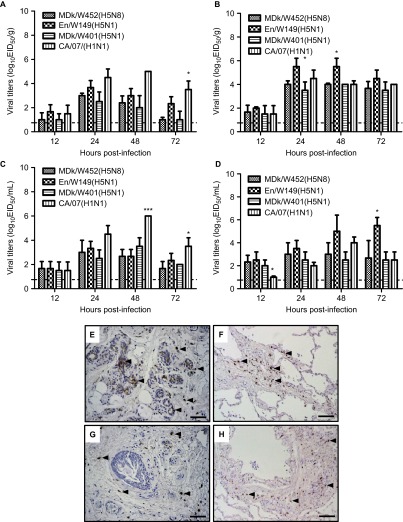
Having documented the novel gene constellation of the 2014 HPAI A(H5N8) virus, we first sought to compare its biological features with those of HPAI A(H5N1) viruses that have been recently isolated in Korea and with those of the 2009 human pandemic H1N1 virus. MDk/W452(H5N8) formed relatively smaller plaques in MDCK cells than did En/W149(H5N1) and MDk/W401(H5N1), previous Korean clade 2.2 and 2.3.2.1 HPAI A(H5N1) isolates, respectively, and exhibited spherical virus particles under transmission electron microscopy (Supplementary Figure S2 and data not shown). Despite the polybasic cleavage motif in HA, MDk/W452(H5N8) inefficiently produced visible plaques in the absence of exogenous protease (Supplementary Figure S2C, right panel). The HPAI A(H5N8) virus grew more rapidly in SPF eggs than did En/W149(H5N1) and MDk/W401(H5N1) (8.9 vs. 8.0–8.3 EID50s) (Supplementary Table S3). However, it could not outgrow En/W149(H5N1), which bears the mammalian-adaptive Glu627Lys substitution in PB2 (PB2627K), in cultured MDCK cells (6.9 vs. 8.6 TCID50s). In in vitro experiments, MDk/W452(H5N8) also had consistently had lower replication titers in differentiated primary CELu (7.5 vs. 9.5 log10TCID50/mL peak titers, P<0.05) (Supplementary Figure S3A) and in NHBE (4.8 vs. 8.5 log10TCID50/mL peak titers, P<0.001) (Supplementary Figure S3D) in trypsin-free media. Despite these differences in cultured cells, MDk/W452(H5N8) replicated in human nasal respiratory epithelium and lung tissues as well as the HPAI A(H5N1) viruses did ex vivo (3.0–4.0 log10EID50/g maximum titers) (Figures 1A–1D). Moreover, it also demonstrated attachment to human respiratory tissues, although this attachment was not as extensive as that exhibited by the control 2009 pandemic CA/07(H1N1) virus (Figures 1E–1F vs. Figures 1G–1H).
Figure 1.
Growth kinetics and attachment of viruses in human respiratory tissues ex vivo. Replication of the MDk/W452(H5N8) virus was monitored in human nasal respiratory epithelial (A) and lung (B) tissue explants and corresponding culture supernatants (C and D, respectively) starting at 12 hpi and at 24-h intervals thereafter. Growth kinetics were compared to those of the control HPAI A(H5N1) (En/W149 and MDk/W401) and 2009 pandemic CA/07(H1N1) viruses. The titers shown are means±SD from three independently performed experiments. Immunohistochemical detection of viral NP antigen was performed using nasal and lung tissue blocks incubated with CA/07(H1N1) (E and F), the control strain, or MDk/W452(H5N8) (G and H), the test virus. Arrows indicate positive immunostaining. Scale bars=50 µm. *P<0.05, ***P<0.0001 relative to MDk/W452(H5N8).
Antiviral compounds represent the first line of therapeutic and prophylactic options in the event of unprecedented human over-spill of the HPAI A(H5N8) virus. Assessment of the IC50 of the NA inhibitors oseltamivir, zanamivir and peramivir against egg-grown virus stocks revealed that MDk/W452(H5N8) was sensitive to this panel of antiviral compounds (Supplementary Table S4), essentially correlating with the absence of drug-resistance markers in the NA gene.
Receptor-binding preference of MDk/W452(H5N8)
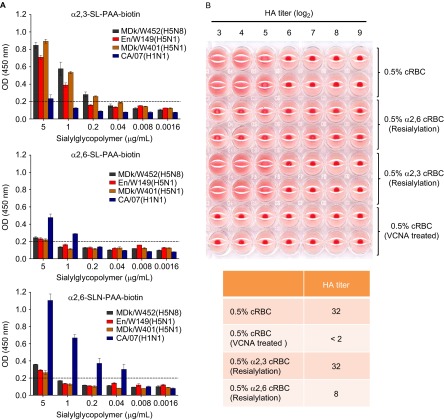
Because the receptor specificity of HA influences virus replication and transmission, we elucidated receptor-binding preference by performing a solid-phase direct binding assay that directly measures the affinity of the virus for polyacrylamide (PAA)-biotin-conjugated glycans Neu5Acα2–3Galβ1–4Glc β1 (α2,3′-SL-PAA-biotin), Neu5Acα2–6Galβ1–4Glc (α2,6′-SL-PAA-biotin) and Neu5Acα2-6Galβ1-4GlcNAc (α2,6′SLN-PAA-biotin).22 The control CA/07(H1N1) virus exhibited a strong binding preference for α2,6-linked sialic acids (α2,6-SAs) (Figure 2A). In contrast, MDk/W452(H5N8) as well as the authentic HPAI A(H5N1) viruses showed a strong binding preference for α2,3′-SL-PAA-biotin residues. Moreover, we also observed binding of MDk/W452(H5N8) to α2,6′-SLN-PAA-biotin probes, which have been proposed to be authentic analogs for influenza virus receptors in mammalian hosts,43 with considerably higher affinity than that displayed by the HPAI A(H5N1) viruses used in this study. Using re-sialyated cRBCs that express either α2,6-SAs or α2,3-SAs,23 we also found that MDk/W452(H5N8) binds α2,6-re-sialylated cRBCs, despite its strong preferential binding to α2,3-resialylated cRBCs (Figure 2B), in agreement with results of our direct virus-binding assays and our data obtained in cultured cells and tissue explants (Figure 1 and Supplementary Figure S3). Thus, the HPAI A(H5N8) virus has strong α2,3-SA receptor-specificity but can also recognize α2,6-SA receptors.
Figure 2.
Virus receptor-binding specificity assays. (A) Binding affinity of inactivated whole viruses to SA α2,3′-SL-PAA-biotin, SA α2,6′-SL-PAA-biotin or SA α2,6′SLN-PAA-biotin glycans. The results shown are means±SD (mean of three replicates). The dashed lines indicate the limit of detection. (B) HA assays using re-sialylated cRBCs. The results obtained with re-sialylated cRBCs were normalized to the results obtained with untreated cRBCs.
Pathogenicity and transmission in domestic poultry
We next compared the virulence of MDk/W452(H5N8), En/W149(H5N1) and MDk/W401(H5N1) in land-based avian poultry species. All HPAI H5 viruses were highly pathogenic in SPF chickens (intravenous pathogenicity indices: 2.74–3.0) (Supplementary Table S3), which exhibited disease signs typical of those infected with HPAI A(H5N1) viruses.44 Virus inoculum was recovered in swabs and detected in the visceral organs tested (Supplementary Table S5 and Table 1).
Table 1. Viral tissue titers in experimentally inoculated animalsa.
| Tissue | MDk/W452(H5N8) | En/W149(H5N1) | MDk/W401(H5N1) | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chicken | Duck | Mouse | Ferret | Chicken | Duck | Mouse | Ferret | Chicken | Duck | Mouse | Ferret | |||||||||||||
| 3db | 5d | 3d | 5d | 3d | 5d | 3d | 5d | 3d | 5d | 3d | 5d | 3d | 5d | 3d | 5d | 3d | 5d | 3d | 5d | 3d | 5d | 3d | 5d | |
| Lung | 6.5 ±0.1c | 5.0±0.2 | 5.0±0.3 | 4.0±0.1 | 2.6±0.3 | 3.0±0.4 | 2.0±0.2 | 2.5±0.3 | 6.0±0.3 | 6.5±0.4 | 6.0±0.3 | 6.5±0.1 | 6.0±0.2 | 6.25±0.2 | 5.0±0.3 | 4.0±0.3 | 5.3±0.1 | 6.0±0.4 | 5.5±0.2 | 6.5±0.3 | 4.8±0.4 | 4.1±0.1 | 3.7±0.1 | 2.5±0.1 |
| Brain | 3.5±0.4 | — | — | — | — | 2 | 1±0.1 | 1.0 | 4.0±0.2 | 4.5±0.2 | 4.0±0.2 | 5.0±0.2 | 3.7±0.3 | 3.0±0.1 | — | — | 3.3±0.2 | 4.0±0.3 | 1.5 | 4.5±0.2 | 3.0±0.2 | 2 | — | — |
| Kidney | 5.5±0.2 | 3.0±0.2 | 3±0.2 | 3.5±0.2 | — | — | — | — | 3.0±0.2 | 3.0±0.2 | 3.5±0.2 | 4.0±0.1 | 1.5±0.1 | 1.0±0.1 | 2.5±0.2 | 1.5 | 1.5±0.1 | 2.0±0.1 | 2.5±0.2 | 3.5±0.2 | 3.5±0.3 | 2.5±0.1 | — | — |
| Spleen | 3.5±0.2 | — | 3.5±0.1 | 1.5 | — | — | — | 1.0±0.1 | 1.5±0.1 | 2.0±0.1 | 3.5±0.1 | 4.5±0.2 | 1.5±0.1 | 1.0±0.1 | 2.0±0.1 | 1 | 2.5±0.2 | 3.0±0.2 | 2.3±0.1 | 4.0±0.1 | 1.0±0.1 | — | — | — |
| Heart | 6.5±0.3 | 2.5±0.1 | 5±0.3 | 2.75±0.1 | — | — | — | — | 2.5±0.1 | 3.0±0.1 | 4.5±0.2 | 5.5±0.3 | 2.5±0.2 | 1.5±0.3 | — | — | 2.5±0.3 | 3.0±0.1 | 3.5±0.2 | 5.0±0.3 | 1.5±0.1 | 1.0±0.2 | — | — |
| Liver | 1.5d | — | 3.5±0.2 | — | — | — | — | 1.5 | 2.5±0.2 | 1.5±0.2 | 4.0±0.2 | 2.5±0.1 | 2.5 | — | — | — | 1.5±0.1 | — | 3.0±0.1 | — | 1.5 | — | — | — |
| Colon | 2.5±0.1 | 2.75±0.2 | 4±0.3 | 3±0.3 | — | — | — | 1.5 | 5.3±0.4 | 5.5±0.3 | 6.5±0.3 | 6.5±0.2 | 2.0±0.1 | 1.0±0.1 | — | — | 5.3±0.4 | 5.5±0.2 | 6.5±0.3 | 6.5±0.3 | 1.5±0.2 | 1.0±0.1 | — | — |
Dashed lines indicate that the tissue was negative for virus detection (lower limit=0.7 log10EID50/g).
Results were obtained from three animals per group at each time point and are expressed as log10 EID50/g tissue.
Day.
Standard deviation titers.
Positive detection in one animal.
By contrast, severe-to-moderate signs of infection were observed in MDk/W452(H5N8)-inoculated ducks. Although the viruses were shed from the oropharynx and cloaca, MDk/W452(H5N8) produced substantially lower oropharyngeal titers than did the A(H5N1) viruses, and the titers persisted only up to 5 dpi (Supplementary Table S5). These results are consistent with results obtained using the parental 2010 Chinese HPAI A(H5N8) virus in mallard ducks.8 Although En/W149(H5N1) and MDk/W401(H5N1) displayed LD50s of 2.0 and 5.3, respectively, in ducks, only approximately 17% of oronasally infected ducks succumbed to infection by the MDk/W452(H5N8) virus (LD50≥6.0) (Supplementary Table S3). Higher levels of virus replication occurred in the lungs, heart, and intestines of MDk/W452(H5N8)-infected ducks sacrificed at 5 dpi than in oropharynx and cloaca, and no virus was detected in brain tissue samples (Table 1 and Supplementary Table S5). Histopathological analysis revealed that lesions in MDk/W452(H5N8)-infected ducks were substantially attenuated and appeared to be less invasive than those in inoculated chickens (Supplementary Figure S4). In contrast to its ready detection in chickens (Supplementary Figures S4A–S4D), viral antigen was rarely detected in the brain and heart muscle of ducks but was readily detectable in intestinal tissues (Supplementary Figures S4E–S4H). All uninfected DC ducks (six of six) harbored the respective HPAI H5 viruses with which they were inoculated (Supplementary Table S3). These data indicate that MDk/W452(H5N8) has relatively low pathogenicity in ducks, unlike in chickens, and does not replicate well or systemically in this species. However, ducks remain susceptible to infection, even by naturally transmitted A(H5N8) virus.
Replication and pathogenesis in mammalian hosts
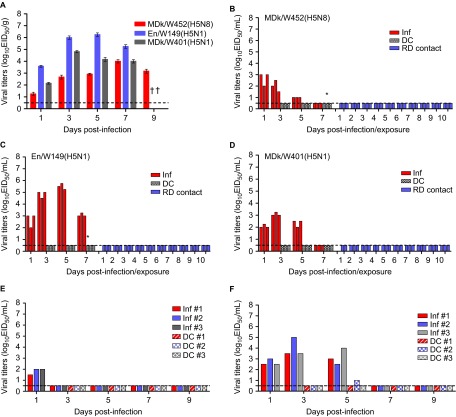
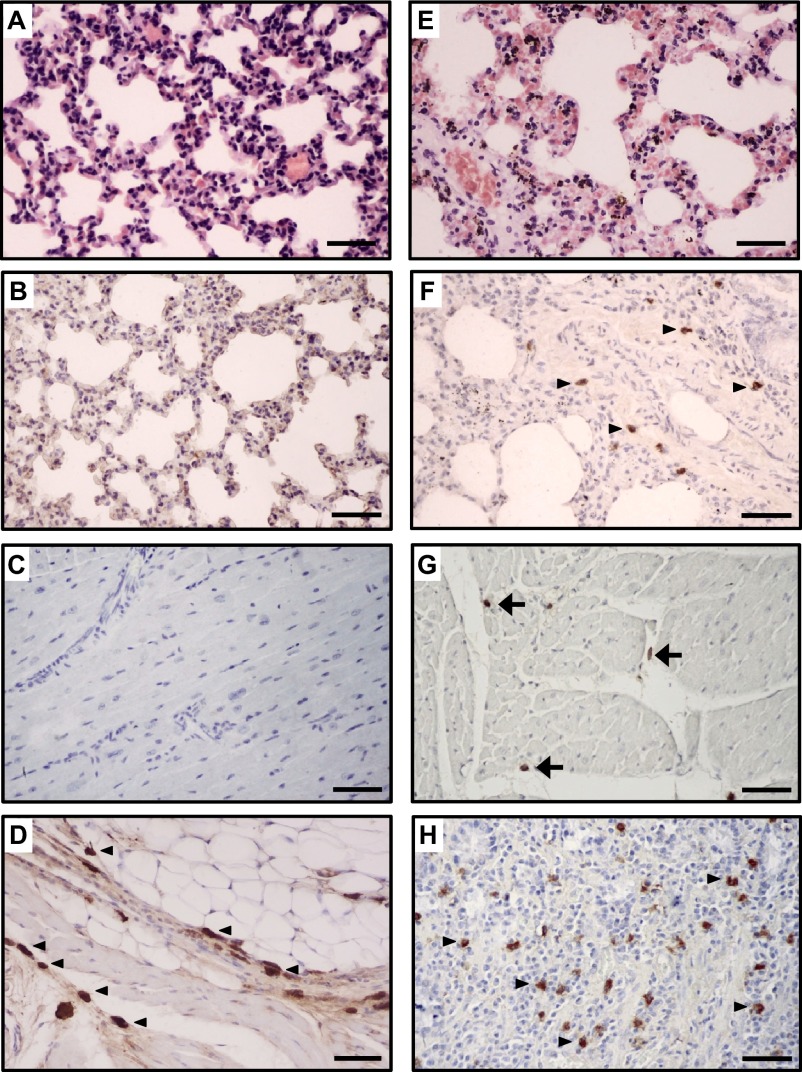
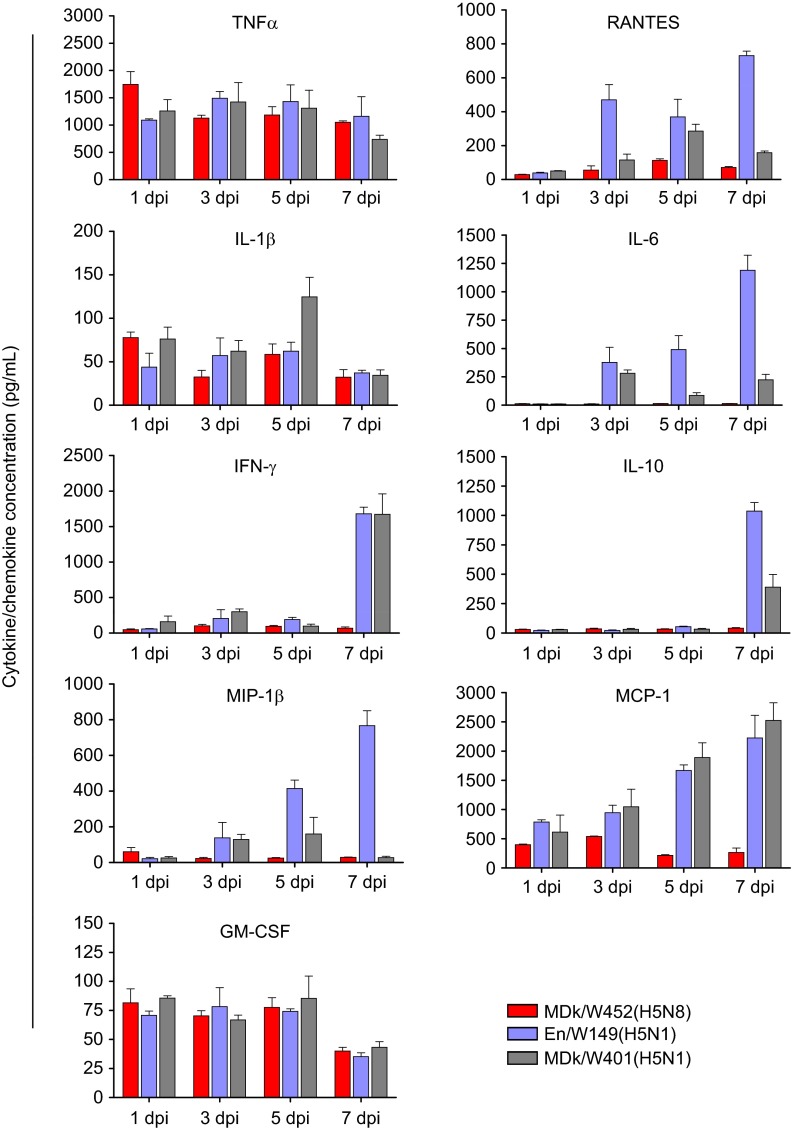
The pathogenicity and pandemic potential of the virus were next examined in mice and ferrets, the established mammalian surrogates for humans in influenza research. In BALB/c mice, MDk/W452(H5N8) was less pathogenic than En/W149(H5N1) and MDk/W401(H5N1), as shown by their 50% mice lethal dose (MLD50) values (5.5, 3.3 and 3.9, respectively) (Supplementary Table S3). In mice inoculated intranasally with 106 EID50/50 µL, MDk/W452(H5N8) induced only 6% reduction in body weight and 40% lethality within the 14-day observation period, whereas En/W149(H5N1) and MDk/W401(H5N1) were very lethal and replicated systemically in various tissues (Supplementary Figure S5). Although detectable up to 9 dpi, the virus replicated moderately in mice lungs (104 EID50/g maximum lung titers) and was only recovered in brain tissues at 5 dpi (Figure 3A and Table 1). Histologically, MDk/W452(H5N8) microscopic lung lesions were not extremely severe and were characterized by mild thickening of the alveolar septa with increased cellularity (Figure 4A). Antigen detection in lung and heart muscle was rare (Figures 4B and 4C) but was abundant in the nasal cavity (Figure 4D). Analysis of mouse BAL fluids indicated that MDk/W452(H5N8) infection upregulated only TNF-α, IL-1β and GM-CSF, whereas the HPAI A(H5N1) viruses, particularly En/W149(H5N1), induced robust expression of all pro-inflammatory cytokines tested (Figure 5). Consistent with this finding, persistent activation and recruitment of immune cells (e.g., T cells, macrophage, monocyte), the hallmark of severely lethal HPAI A(H5N1) virus infection, was not observed with A(H5N8). Thus, in mice, MDk/W452(H5N8) was moderately pathogenic relative to the HPAI A(H5N1) viruses.
Figure 3.
Replication of MDk/W452(H5N8) in mammalian animal models. Virus replication was examined in mice, ferrets, dogs and cats that had been experimentally inoculated intranasally with 106 EID50/mL of designated virus. Mouse lung titers shown are means±SD from three animals (A). Individual nasal wash titers are shown for ferrets inoculated with MDk/W452(H5N8) (B), En/W149(H5N1) (C) or MDk/W401(H5N1) (D). To examine transmission, the inoculated animals were individually paired with a DC and an RD-contact animal (1∶1∶1 setup, triplicate). Three infected dogs (E) and cats (F) were also individually cohoused with contact animals in the same cage at 1 dpi and monitored for virus shedding in nasal swabs. The limit of virus detection was 0.7 log10EID50/mL; titers below that limit are shown as 0.5 log10EID50/mL (dashed lines). An asterisk (*) signifies seroconversion as determined by HI analysis at 21 dpi. Inf, directly infected animal.
Figure 4.
Histopathology and immunohistochemical staining for influenza virus antigen in tissues of mouse (A–D) and ferret (E–H) infected with the HPAI A(H5N8) virus. In the lung, the alveolar septum is mildly thickened with increased cellularity (mouse: A and B; ferret: E and F). The influenza viral NP antigen was rarely detected in the lungs and hearts of mice (B and C, respectively) and was infrequently detected in mononuclear phagocytic cells in the lungs and hearts of ferrets (F, arrows; G, arrowheads, respectively). In contrast, in the nasal cavities of mice (D, arrows) and in the intestines of ferrets (H), mononuclear phagocytic cells were commonly immunopositive for influenza virus antigen. Scale bars=50 µm.
Figure 5.
Cytokine and chemokine responses in the lungs of infected mice. Concentrations of various cytokines/chemokines in BAL fluids of infected mice at 1, 3, 5 and 7 dpi were measured by protein analysis with the Luminex-based multiplex immunoassay kit (Affymetrix, Santa Clara, CA, USA). The values shown are means±SD (errors bars) from three mouse BAL fluids per time point tested.
In influenza-free ferrets, MDk/W452(H5N8) transiently elevated body temperature without producing any remarkable signs of illness after intranasal inoculation (106 EID50/mL) (Supplementary Figure S6A). In contrast, infection with En/W149(H5N1) reduced food consumption, resulting in 10% maximum weight loss and reduced activity (Supplementary Figure S6B). All three HPAI H5 viruses were shed from the upper respiratory tract, but MDk/W452(H5N8)-infected ferrets consistently produced lower nasal wash virus titers that were detectable only up to 3 dpi (Figures 3B–3D). Despite the shortened period of time and lower titer of virus excretion, MDk/W452(H5N8) replicated in the lungs and spleen and was the only HPAI H5 virus that was also recovered from brain, liver, and intestine (Table 1). As in mice, moderate histopathological lesions were observed in ferret lungs, accompanied by infrequent detection of antigen-positive cells in the heart and its abundant presence in intestinal tissues (Figures 4E–4H). In comparison, the En/W149(H5N1) virus was also detected immunohistologically in the brain (Supplementary Figures S7A–S7D vs. Supplementary Figures S7E–S7H).
To determine transmissibility, each of the inoculated animals was individually cohoused with a naïve DC ferret at 1 dpi; a RD contact was also placed in the same isolator adjacent to a cage containing an inoculated ferret to assess aerosol transmission (1∶1∶1 set-up in triplicate). RD-contact ferrets were separated by stainless steel grill dividers that only permitted virus dissemination through the air. Virus was not recovered in the nasal washes of any of the DC- or RD-contact ferrets exposed to the HPAI H5 viruses (Figures 3B–3D, right panels). However, one of three DC ferrets exposed to MDk/W452(H5N8)-infected or En/W149(H5N1)-infected animals demonstrated seroconversion in HI assays performed at 21 dpi (40 HI units), as did directly inoculated animals, indicating inefficient contact transmission (Supplementary Table S6). Because replication and transmission of animal viruses in ferrets are sometimes associated with amino acid changes in the viral genome,16,34,45 we sequenced the viruses recovered from the nasal washes and tissues of inoculated animals. Compared to the sequence of the virus inoculum, the sequences of these samples contained one non-synonymous mutation in the HA2 region of the H5 HA (Lys480Asn, H3 numbering) and one in the PB2 region (Glu701Asn) (data not shown). ThePB2701N substitution harbored by the virus recovered from the lung and intestine of an inoculated ferret sacrificed at 5 dpi is a known mammalian host-adaptation and virulence marker.38
Dogs and cats are moderately susceptible to MDk/W452(H5N8)
On several farms, dogs developed antibodies against the A(H5N8) virus, prompting our investigation in this host. MDk/W452(H5N8) did not induce observable signs of illness in dogs, consistent with a lack of efficient virus replication in the upper nasal cavity and visceral tissues of this species (Figure 3E and Supplementary Figures S8A–S8C). Co-housed DC dogs did not shed the virus, and only directly inoculated dogs seroconverted at the end of the experiment (Supplementary Table S7). In contrast to beagles, domestic cats had transient fever and showed marginal weight loss (one of three) after inoculation (Supplementary Figures S8D–S8E). MDk/W452(H5N8) was shed in nasal swabs up to 5 dpi, and appreciable virus titers were also detected in the nasal turbinates, trachea and lung tissue samples (Figure 3F and Supplementary Figure S8F). One of three DC cats was tentatively positive for virus detection 4 days after exposure, but did not sustain virus replication (Figure 3F, right panel). As in dogs, cats that were directly inoculated developed antibodies against A(H5N8) at low HI titers (20–40 units), but cats that were only exposed to directly inoculated animals did not (Supplementary Table S7). In inoculated dogs and cats, pneumonitis was observed in the lungs, and the lesions appeared to increase at 5 dpi (Supplementary Figure S9). Multiple consolidated areas were composed mainly of macrophages and pneumocytes, and antigen-positive alveolar macrophages were observed in both animal models. These data suggest that the novel A(H5N8) virus can replicate in the respiratory tissues of directly inoculated canine and feline hosts, with cats being more susceptible than dogs to infection. However, transmission by DC was not established. Therefore, although the seroconversion observed in dogs indicates potential interspecies transmission in the field, our results suggest that extensive close contact with infected poultry may be required for positive infection.
DISCUSSION
Since the incursion of the current HPAI A(H5N8) virus in domestic poultry, at least 12 million avian poultry have been culled, the largest number destroyed since 2003. In wild birds, the virus has been detected virologically and serologically in various species including teals (Baikal and green-winged), ducks (mallard and spot-billed), geese (bean and white-fronted), whooper swans and egrets. Thus, our isolation of MDk/W452(H5N8) in the mid-region of Korea just a few weeks after its first detection in the southwestern Jeolla provinces stresses the major role of wild migratory waterfowl in the spread of the virus. Consistent with this, Japan also reported similar poultry outbreaks due to the HPAI A(H5N8) virus in April 2014.46
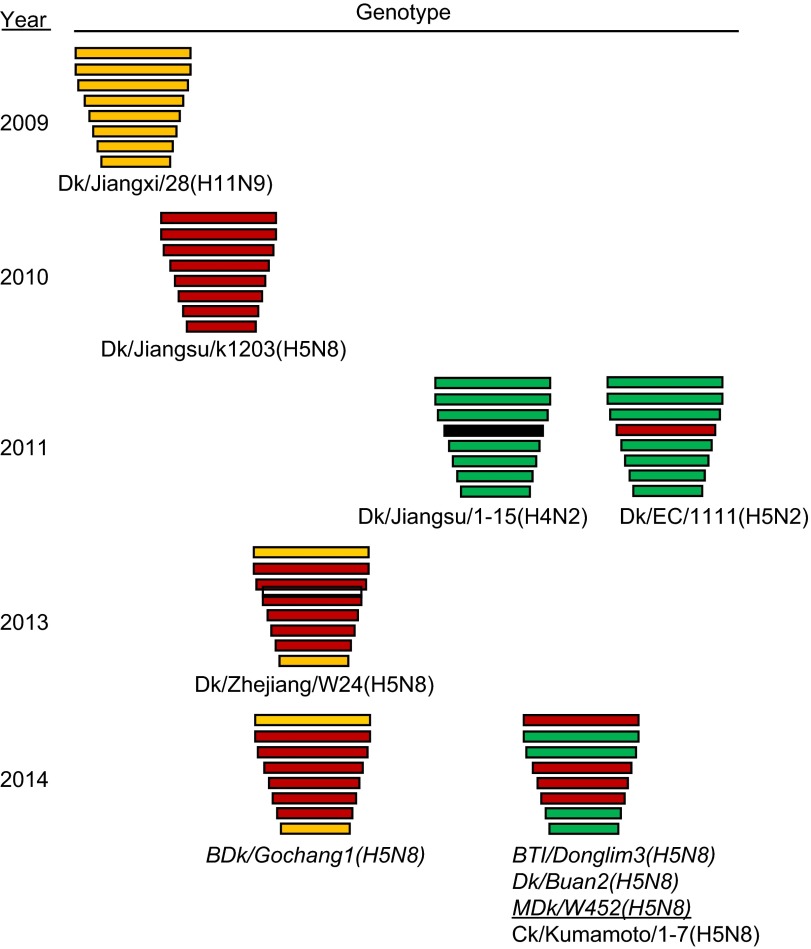
HPAI A(H5N1) viruses have been discretely involved in reassortment events that have either generated H5 strains with novel NA subtypes or donated gene components to other co-circulating strains.3,4,5,6,7,8,9,47 In Asia, A(H5N8) viruses have been found in China, Hong Kong, Thailand, and Korea, but none of those previous strains resembled the genetic components of this 2014 viral pathogen nor did they cause local outbreaks. Because China, Korea and Japan share interconnected wild bird migratory routes,48 it is not surprising that the novel HPAI A(H5N8) virus would be shared by these eastern Asian nations. A very recent report described the isolation of A(H5N8) viruses in China in December 2013 that are genotypically identical to BDk/Gochang1(H5N8),49 a reassortant between HPAI A(H5N8)-like and A(H11N9)-like strains.15 The timeline of their detection suggests that BDk/Gochang1(H5N8) likely descended from those 2013 Chinese isolates (Figure 6). Less is known about the emergence of the MDk/W452(H5N8)-like viruses, which appeared to have been predominantly isolated in Korea and subsequently carried to Japan by migratory birds. It is unclear which HPAI A(H5N8) virus lineage (the 2010 Jiangsu-like or 2013 Zhejiang-like strain) participated in reassortment. Regardless, the available data indicate that HPAI A(H5N8)-like viruses underwent independent reassortment events with other viral strains found in wild birds in eastern China (e.g., H11N9, H4N2 and/or H5N2), emphasizing the role of migratory waterfowl in the creation, perpetuation, and spread of newly emerging avian viruses (Figure 6). One notable feature of the novel 2014 MDk/W452(H5N8) virus is the C-terminal amino acid extension of its NS1 protein, a feature that is not present in any of the parental viruses and is indicative of further genetic evolution. This C-terminal extension, a genetic feature among human viruses prevalent from 1950 to 1987, creates a functional nucleolar localization signal that is thought to affect pathogenesis.50 However, the genetic impact of this feature on the newly emergent HPAI A(H5N8) virus is currently unknown.
Figure 6.
Putative generation of the novel HPAI A(H5N8) viruses that caused domestic poultry outbreaks in Korea and Japan in 2014. Genes from top to bottom are PB2, PB1, PA, HA, NP, NA, M and NS. For simplicity, each color represents an individual virus background. Italicized virus names are the Korean strains, and the underlined virus is the isolate we characterized. Virus abbreviations: Dk/Jiangxi/28(H11N9), A/duck/Jiangxi/28/2009(H11N9); Dk/Jiangsu/k1203(H5N8), A/duck/Jiangsu/k1203/2009(H5N8); Dk/Jiangsu/1-15(H4N2), A/duck/Jiangsu/1-15/2011(H4N2); Dk/EC/1111(H4N2), A/duck/Eastern China/1111/2011(H4N2); Dk/Zhejiang/W24(H5N8), A/duck/Zhejiang/W24/2013(H5N8); BDk/Gochang1(H5N8), A/broiler duck/Korea/Gochang1/2014(H5N8; BTl/Donglim3(H5N8), A/Baikal teal/Korea/Donglim3/2014(H5N8); Dk/Buan2(H5N8), A/duck/Korea/Buan2/2014(H5N8); MDk/W452(H5N8), A/mallard duck/W452/2014(H5N8); Ck/Kumamoto/1-7(H5N8), A/chicken/Kumamoto/1-7/2014(H5N8).
In domestic poultry, MDk/W452(H5N8) was shed in high titers, and it replicated systemically in chickens, retaining its high-pathogenicity phenotype. However, this virulent pathotype did not extend to domestic ducks despite their vulnerability to infection. The rapid excretion of this virus coupled with the limited morbidity it produces could present a significant challenge to virus monitoring efforts on domestic farms. If poultry owners rely mainly on mortality (due to exacerbated disease) or reduced egg production as visible criteria for detection, the virus could spread long before infection is suspected or confirmed. Hence, apart from migratory wild birds, the movement of asymptomatic ducks may have also contributed to the silent spread of the virus that gave rise to the recent unprecedented and widespread poultry outbreaks. Furthermore, the results suggest that MDk/W452(H5N8) has the potential to become established in domestic ducks and to alter the genetic evolution of pre-existing avian viruses in poultry.
Multiple viral genetic factors contribute to the pathogenesis and transmission of influenza viruses. Although these characteristics are often linked to viral surface glycoproteins, internal gene segments also contribute to virus adaptation, pathogenesis, transmission, and immune evasion. Among these segments, PB2 with the accompanying 627K mutation has been frequently implicated in many of these aspects.38 On one hand, the existence of such a factor might help explain why MDk/W452(H5N8), which lacks the PB2627K marker, displayed relatively impaired replicative ability and inefficient contact transmission in our mammalian models despite the fact that it harbors a polybasic HA cleavage site and PB1-F266S molecular determinants. On the other hand, the novel gene constellation of the 2014 HPAI A(H5N8) virus may not yet be optimized so as to facilitate growth in mammalian hosts. However, reconstructed H5 viruses require only a few mutations to become capable of efficient transmission in ferrets.34,35,45 Thus, it is interesting to note that the host-adaptive PB2701N mutation was acquired non-synonymously during replication in a ferret host, indicating the potential of the virus to evolve genetically. At the same time, the genetic repercussion of the HA mutation located at the relatively stable HA2 molecule, if any, remains to be elucidated. Regardless, these genetic substitutions could indicate that continuous circulation and unprecedented replication in mammals might provide a means by which the A(H5N8) virus can acquire important molecular determinants that could alter host range, replication, virulence, and/or transmission in appropriate hosts. Additionally, the observed susceptibility of dogs and cats could provide alternative intermediate hosts for virus evolution. However, we maintain that extensive close contact with affected poultry remains a significant requirement for interspecies transmission to occur.
In summary, we show that prevailing eastern Asian HPAI H5 viruses actively interact with cocirculating avian strains in the field through genetic reassortment. These events may help set the stage for the generation of novel viruses that could pose future human health concerns. Nevertheless, our results reinforce the initial impression that there is no imminent threat from the newly emergent 2014 HPAI A(H5N8) virus. However, the observed binding of this viral strain to mammalian-like receptors and its ability to replicate in mammalian models, albeit at moderate levels, indicate that its evolution into a notable virus remains a possibility that should not be ignored. Avian influenza viruses circulating in poultry capable of directly infecting humans can emerge, as exemplified by various A(H5N1) and, most recently, A(H7N9) viruses. Fortunately, some available prophylactic compounds are helpful in alleviating the clinical disease burden if human infections occur in the absence of appropriate vaccines. However, because the 2014 HPAI A(H5N8) virus may have the potential to become endemic in domestic ducks, which serve as an interface between the natural gene pool of avian influenza viruses in wild birds and land-based poultry, persistent monitoring and systematic surveillance should be maintained for containment purposes and to reduce opportunities for further genetic evolution of the virus.
Acknowledgments
We thank Dr Cherisse Guess (St Jude Children's Research Hospital, Memphis, TN, USA) for her excellent editorial assistance. This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science (NRF-2007-0054930) and by the Korea Healthcare Technology R&D Project funded by the Ministry of Health (Grant NO A103001). This project was also funded in part (to Richard J Webby and Robert G Webster) with federal funds from the Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under Centers of Excellence for Influenza Research and Surveillance (contract number HHSN272201400006C) and by the American Lebanese-Syrian Associated Charities.
Footnotes
Note: Supplementary Information for this article can be found on Emerging Microbes and Infections' website (http://www.nature.com/emi/).
Supplementary Information
References
- Eagles D, Siregar ES, Dung DH, Weaver J, Wong F, Daniels P. H5N1 highly pathogenic avian influenza in Southeast Asia. Rev Sci Tech. 2009;28:341–348. doi: 10.20506/rst.28.1.1864. [DOI] [PubMed] [Google Scholar]
- World Health Organization/Global Influenza Programme Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2014 Geneva: WHO; 2014. Available at http://www.who.int/influenza/human_animal_interface/EN_GIP_20140124CumulativeNumberH5N1cases.pdf?ua=1 (accessed 24 January 2014). [Google Scholar]
- Chen H, Smith GJ, Li KS, et al. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc Natl Acad Sci USA. 2006;103:2845–2450. doi: 10.1073/pnas.0511120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Liu W, Cao Y, et al. Novel reassortant highly pathogenic avian influenza (H5N5) viruses in domestic ducks, China. Emerg Infect Dis. 2011;17:1060–1063. doi: 10.3201/eid1706.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Huang J, Chen Y, et al. Genome sequence of a natural reassortant H5N2 avian influenza virus from domestic mallard ducks in eastern China. J Virol. 2012;86:12463–12464. doi: 10.1128/JVI.02315-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Peiris JS, Lipatov AS, et al. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc Natl Acad Sci USA. 2002;99:8950–5955. doi: 10.1073/pnas.132268999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Gu X, Lu X, et al. Novel reassortant highly pathogenic H5N2 avian influenza viruses in poultry in China. PLoS One. 2012;7:e46183. doi: 10.1371/journal.pone.0046183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Gu M, Zhong L, et al. Characterization of three H5N5 and one H5N8 highly pathogenic avian influenza viruses in China. Vet Microbiol. 2013;163:351–357. doi: 10.1016/j.vetmic.2012.12.025. [DOI] [PubMed] [Google Scholar]
- Zou W, Guo X, Li S, Yang Y, Jin M. Complete genome sequence of a novel natural recombinant H5N5 influenza virus from ducks in central China. J Virol. 2012;86:13878. doi: 10.1128/JVI.02725-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YK, Joh SJ, Kim MC, et al. Highly pathogenic avian influenza (H5N1) in the commercial domestic ducks of South Korea. Avian Pathol. 2005;34:367–370. doi: 10.1080/03079450500181257. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Choi YK, Kim YJ, et al. Highly pathogenic avian influenza virus (H5N1) in domestic poultry and relationship with migratory birds, South Korea. Emerg Infect Dis. 2008;14:487–490. doi: 10.3201/eid1403.070767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HR, Park CK, Lee YJ, et al. An outbreak of highly pathogenic H5N1 avian influenza in Korea, 2008. Vet Microbiol. 2010;141:362–366. doi: 10.1016/j.vetmic.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Kim HR, Lee YJ, Park CK, et al. Highly pathogenic avian influenza (H5N1) outbreaks in wild birds and poultry, South Korea. Emerg Infect Dis. 2012;18:480–483. doi: 10.3201/eid1803.111490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Organization for Animal Health/World Animal Health Information Database WAHID weekly disease information. Vol. 20, No. 04, 17 January 2014 Paris: OIE; 2012. Available at http://www.oie.int/wahis_2/public/wahid.php/Diseaseinformation/WI (accessed 4 June 2014). [Google Scholar]
- Lee YJ, Kang HM, Lee EK, et al. Novel reassortant influenza A(H5N8) viruses, South Korea, 2014. Emerg Infect Dis. 2014;20:1087–1089. doi: 10.3201/eid2006.140233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascua PN, Song MS, Lee JH, et al. Virulence and transmissibility of H1N2 influenza virus in ferrets imply the continuing threat of triple-reassortant swine viruses. Proc Natl Acad Sci USA. 2012;109:15900–15905. doi: 10.1073/pnas.1205576109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukert PD. Immunofluorescence of avian infectious bronchitis virus in primary chicken embryo kidney, liver, lung, and fibroblast cell cultures. Arch Gesamte Virusforsch. 1966;19:265–272. doi: 10.1007/BF01241849. [DOI] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- Pascua PN, Lim GJ, Kwon HI, et al. Emergence of H3N2pM-like and novel reassortant H3N1 swine viruses possessing segments derived from the A (H1N1) pdm09 influenza virus, Korea. Influenza Other Respir Viruses. 2013;7:1283–1291. doi: 10.1111/irv.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar A. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol Biol. 2000;132:221–241. doi: 10.1385/1-59259-192-2:221. [DOI] [PubMed] [Google Scholar]
- Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Matrosovich MN, Gambaryan AS. Solid-phase assays of receptor-binding specificity. Methods Mol Biol. 2012;865:71–94. doi: 10.1007/978-1-61779-621-0_5. [DOI] [PubMed] [Google Scholar]
- Paulson JC, Rogers GN. Resialylated erythrocytes for assessment of the specificity of sialyloligosaccharide binding proteins. Methods Enzymol. 1987;138:162–168. doi: 10.1016/0076-6879(87)38013-9. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Shinya K, Kiso M, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–1025. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, de Wit E, van den Brand JM, et al. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science. 2009;325:481–483. doi: 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shi J, Deng G, et al. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science. 2013;341:410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Kiso M, Fukuyama S, et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature. 2013;501:551–555. doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xiao H, Lei F, et al. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science. 2005;309:1206. doi: 10.1126/science.1115273. [DOI] [PubMed] [Google Scholar]
- Gu M, Zhao G, Zhao K, et al. Novel variants of clade 2.3.4 highly pathogenic avian influenza A(H5N1) viruses, China. Emerg Infect Dis. 2013;19:2021–2024. doi: 10.3201/eid1912.130340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Li Q, Zhong L, et al. Complete genomic sequence of a novel reassortant H4N2 avian influenza virus isolated from domestic ducks in Jiangsu, China. Genome Announc. 2013;1:e0009113. doi: 10.1128/genomeA.00091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yuan H, Gao R, et al. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster M, van Boheemen S, de Graaf M, et al. Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell. 2014;157:329–339. doi: 10.1016/j.cell.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007;3:1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley JL, Hornung F, Boyd KL, et al. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe. 2007;2:240–249. doi: 10.1016/j.chom.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina RA, Garcia-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Doudna JA. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci USA. 2009;106:21312–21316. doi: 10.1073/pnas.0911915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao P, Tian G, Li Y, et al. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol. 2008;82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intharathep P, Laohpongspaisan C, Rungrotmongkol T, et al. How amantadine and rimantadine inhibit proton transport in the M2 protein channel. J Mol Graph Model. 2008;27:342–348. doi: 10.1016/j.jmgm.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Orozovic G, Orozovic K, Lennerstrand J, Olsen B. Detection of resistance mutations to antivirals oseltamivir and zanamivir in avian influenza A viruses isolated from wild birds. PLoS One. 2011;6:e16028. doi: 10.1371/journal.pone.0016028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Sabesan S, Skehel JJ, Wiley DC. Binding of the influenza A virus to cell-surface receptors: structures of five hemagglutinin–sialyloligosaccharide complexes determined by X-ray crystallography. Virology. 1997;232:19–31. doi: 10.1006/viro.1997.8526. [DOI] [PubMed] [Google Scholar]
- Suarez DL, Perdue ML, Cox N, et al. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J Virol. 1998;72:6678–6688. doi: 10.1128/jvi.72.8.6678-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfst S, Schrauwen EJ, Linster M, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Organization for Animal Health/World Animal Health Information Database WAHID weekly disease information. Vol. 27, No. 16, 13 April 2014 Paris: OIE; 2012. Available at http://www.oie.int/wahis_2/public/wahid.php/Diseaseinformation/WI (accessed 4 June 2014). [Google Scholar]
- Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnberg S, Webby RJ, Webster RG. Natural history of highly pathogenic avian influenza H5N1. Virus Res. 2013;178:63–77. doi: 10.1016/j.virusres.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Peng X, Xu L, et al. Novel reassortant influenza A(H5N8) viruses in domestic ducks, eastern China. Emerg Infect Dis. 2014;20:1315–1318. doi: 10.3201/eid2008.140339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen K, Kinnunen L, Fagerlund R, et al. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J Virol. 2007;81:5995–6006. doi: 10.1128/JVI.01714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.