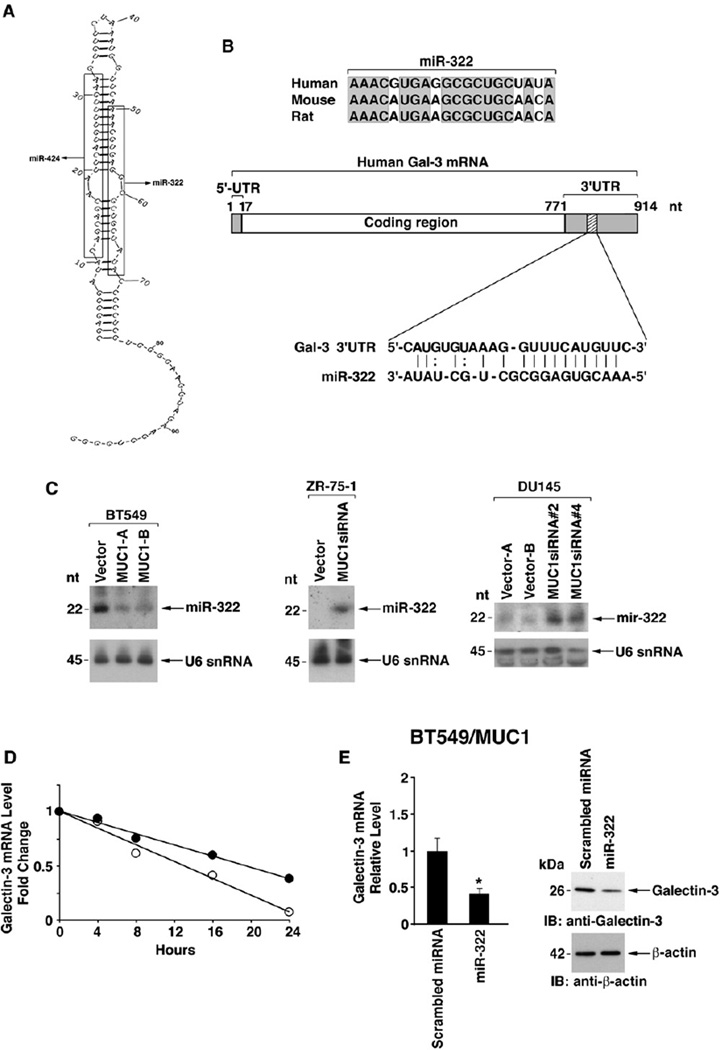
Figure 3. MUC1 Downregulates MiR-322 and Thereby Increases Stability of Galectin-3 mRNA.
(A) Model of the pre-miR stem-loop structure encoding miR-424 and miR-322.
(B) Sequence alignment of human miR-322 with mouse and rat miR-322 and with the galectin-3 3′UTR.
(C) Northern blot analyses of RNA from the indicated BT549, ZR-75-1, and DU145 cells probed for miR-322 (upper panels) and U6 snRNA as a loading control (lower panels).
(D) BT549/vector cells were transfected with an antisense 2′-O-methyl oligoribonucleotide targeted against miR-322 or a scrambled 2′-O-methyl oligoribonucleotide as a control for 48 hr. The cells were treated with actinomycin D and harvested at the indicated times. Total RNA was analyzed for galectin-3 and GAPDH mRNA levels by quantitative RT-PCR. The results are expressed as relative galectin-3 mRNA levels for BT549 cells transfected with the scrambled oligo (○) or the anti-miR-322 (●).
(E) BT549/MUC1 cells were transfected with a pre-miR-322 or a scrambled miRNA and selected in the presence of blasticidin. Cells were assayed for galectin-3 mRNA (left) and protein (right). The results (mean±SD for three replicates) are expressed as the relative galectin-3 mRNA levels (normalized to GAPDH) compared to that obtained with the scrambled miRNA (assigned a value of 1) (left). The asterisk (*) denotes a significant difference at p < 0.01 as compared to control (left).