Fig. 1.
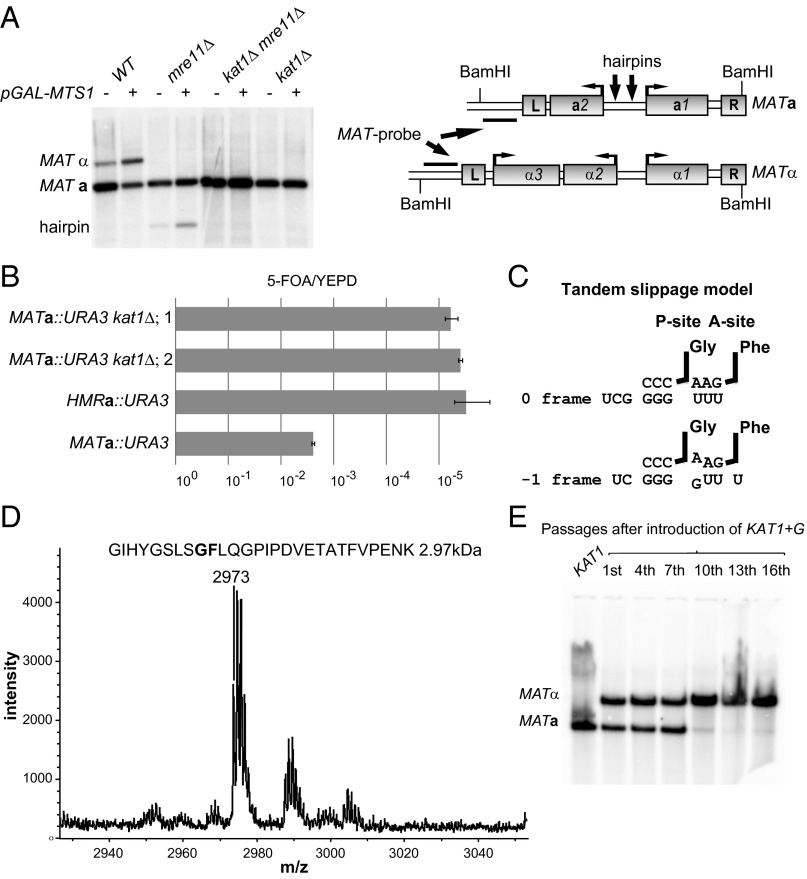
Kat1 is essential for mating-type switching and is subjected to a programmed −1 frameshift. (A, Left) DNA blot analysis of BamHI-digested genomic DNA from WT, mre11Δ, kat1Δ, and mre11Δ kat1Δ containing plasmid alone (−) or pGAL–MTS1 (+), hybridized with a MAT-specific probe. The MATa, MATα, and hairpin bands are indicated. (Right) Schematic diagram of the MATa and MATα loci. Boxes denote genes and the Left (L) and Right (R) repetitive elements. The probe used, the BamHI sites, and approximate positions of hairpins (arrows) are indicated. (B) The ratios of 5-FOA/yeast extract peptone dextrose (YEPD) plating efficiencies were determined in MATa::URA3, HMRa::URA3, and two independently generated MATa::URA3 kat1Δ strains. Error bars represent the SEM from three independent measurements. (C) Tandem slippage model for frameshifting at the KAT1 slippery site. The ribosomal P- and A-sites harboring the tRNAGly and tRNAPhe are indicated. The zero-frame and −1 frame base pairings between the tRNAs and mRNA are shown. (D) MS analysis of a peptide obtained from a GST–Kat1 slippery site–MBP fusion protein. The predicted peptide (2.97 kDa) from the tandem-slippage hypothesis is shown above the graph. The x axis shows the molecular weight in daltons. (E, Lower) DNA blot analysis as described in A. DNA was prepared from a strain carrying the KAT1+G allele at the endogenous locus. (Upper) The number of overnight passages in synthetic complete medium after the generation of the KAT1+G strain.