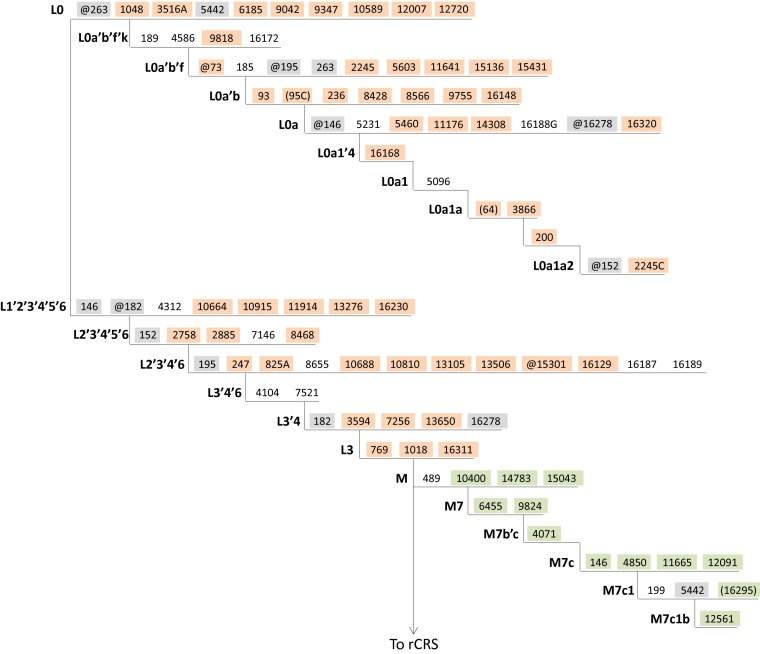
Fig. 1.
The figure displays a simplified representation of the revised Cambridge Reference Sequence (rCRS)-oriented version of the currently accepted human mtDNA phylogeny (data from PhyloTree Build 16, ref. 2) that includes only the branches relevant for sample HG01108. Symbols and nomenclature are consistent with PhyloTree, where the “@” symbol signifies mutation toward the rCRS state, nucleotide positions in parentheses represent mutations that may or may not be present, and inclusion of a nucleotide after the base position indicates a transversion. The presence or absence of the mutations highlighted in this figure among the heteroplasmies reported for sample HG01108 provide evidence for a mixture of two distinct individuals. Sample HG01108 heteroplasmies that can be ascribed to haplogroup L0a1a2 are highlighted in orange and sample HG01108 heteroplasmies at positions diagnostic for haplogroup M7c1b are highlighted in green. Branch positions highlighted in gray represent a mutation and reversion combination on the path between the L0a1a2 and M7c1b lineages (positions 146, 152, 182, 195, 263, and 16278), or homoplasy (position 5442), and thus would not be expected to be observed as variant in a mixture of individuals of these two ancestries (and were not reported as heteroplasmic for sample HG01108). L0a1a2 and M7c1b haplogroup diagnostic positions that were not reported as heteroplasmic in sample HG01108 (positions not highlighted) may be accounted for by (i) the 2,930 mtDNA positions that failed quality control standards and thus were not examined by Ye et al. (1) for any sample, and (ii) additional potentially variant positions on a by-sample basis that did not meet the authors’ criteria for heteroplasmy designation (in addition to less-likely explanations, such as reversion as a private mutation). For the seven heteroplasmies reported for sample HG01108 that were not ascribed to either haplogroup (at positions 5112, 12616, 12684, 13095, 15891, 16362, and 16519), these may be due to either (i) private mutations in either individual represented in the mixture or (ii) true mtDNA heteroplasmy.