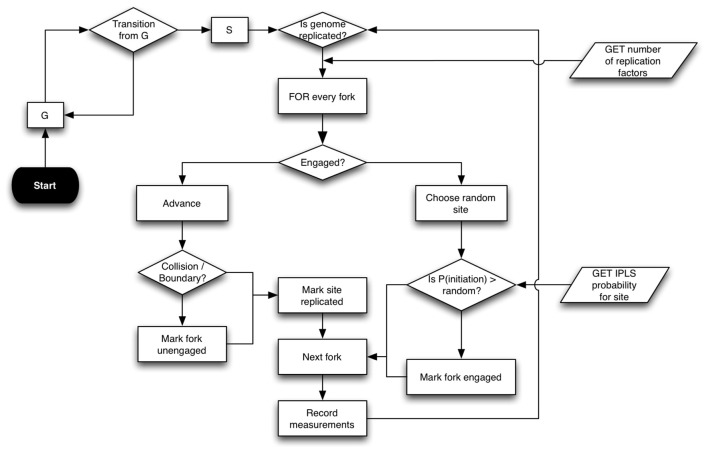
Figure 1.
Replicon algorithm flowchart. Replicon predicts DNA replication timing by simulating cell-cycles in an asynchronous cell population. A simulated cell exists in either G (resting) or S (synthesis) state. While G to S transition occurs at random, the transition from S to G occurs upon completion of genome replication. Upon entering the S state, each cell queries the status of replication forks at its disposal. Forks that are engaged in replication are advanced by an interval (governed by IPLS resolution). Forks are disengaged if their advancement causes either a collision with another fork or if the fork reaches chromosome boundary. If the cell has at least two un-engaged forks at its disposal, then Replicon chooses a random unreplicated chromosome position and initiates replication with probability specified for that position in the IPLS. Replication then proceeds bidirectionally.