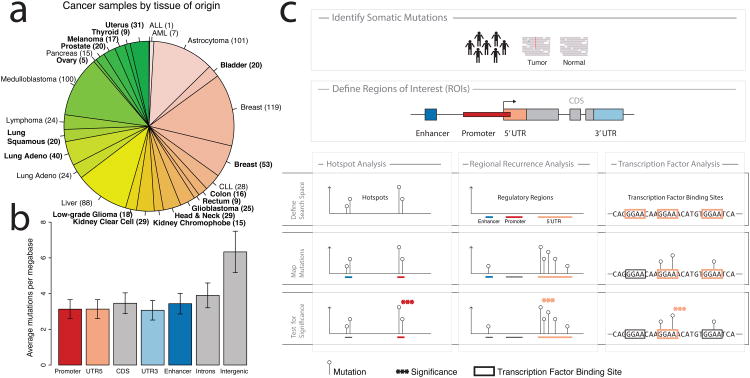
Figure 1.
a) Tumor samples by disease type. Tumor types from TCGA are labeled in boldface; other published samples15 are shown in regular font. b) Mean mutation frequency and 95% confidence interval (n=858) across samples by type of genomic region. c) Workflow for identification of recurrent, non-coding mutations in regulatory regions of interest. Our approach integrates mutation calls from 863 tumor/normal pairs and regulatory regions of interest (ROIs), which are tested for non-coding mutations using three distinct analyses. Hotspot analysis detects recurrent mutations that are often very focal. Regional recurrence analysis identifies annotated regions of interest that are enriched for mutation throughout the entire region. Transcription factor analysis searches for regions that contain recurrent mutations within transcription factor binding sites.