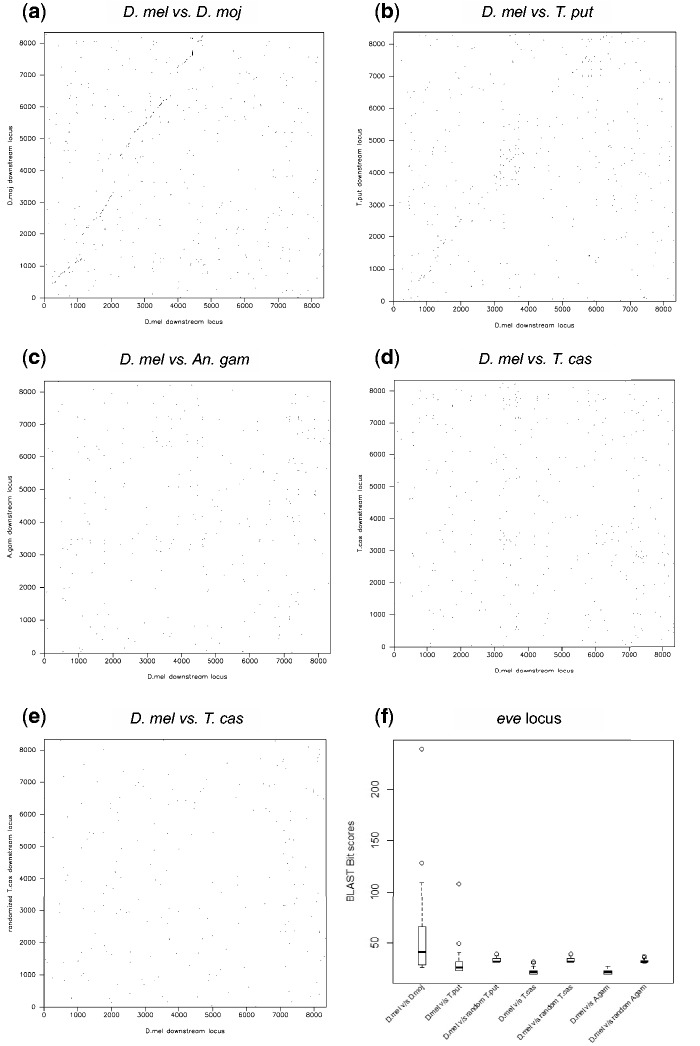
Fig. 3.—
Alignment of intergenic regions of developmental genes between diverged species. Examples in this figure are from the even skipped (eve) locus; see supplementary figure S1, Supplementary Material online, for examples from other loci. (a) Dotplot alignment of Drosophila mojavensis and D. melanogaster (∼60 Ma) downstream intergenic regions shows clear alignment (diagonal) between the two species. (b) The more diverged Sepsid fly Themira putris (∼75 Ma) lies near the edge of noncoding alignment (weak diagonal) as previously reported by Crocker and Erives (2008) and Hare et al. (2008). Neither mosquitoes (c) nor beetles (d) show recognizable alignment, as can be seen from the absence of a visible diagonal and similarity to alignment with randomized sequence (e). The dotplot results are confirmed by BLAST analysis (f), which shows that only D. mojavensis and T. putris have a BLAST score distribution with scores exceeding those obtained from randomized sequence (for D. mel–T. putris vs. D. mel–randomized T. putris, P < 0.024, uncorrected one-sided Student’s t-test).