Fig. 2.—
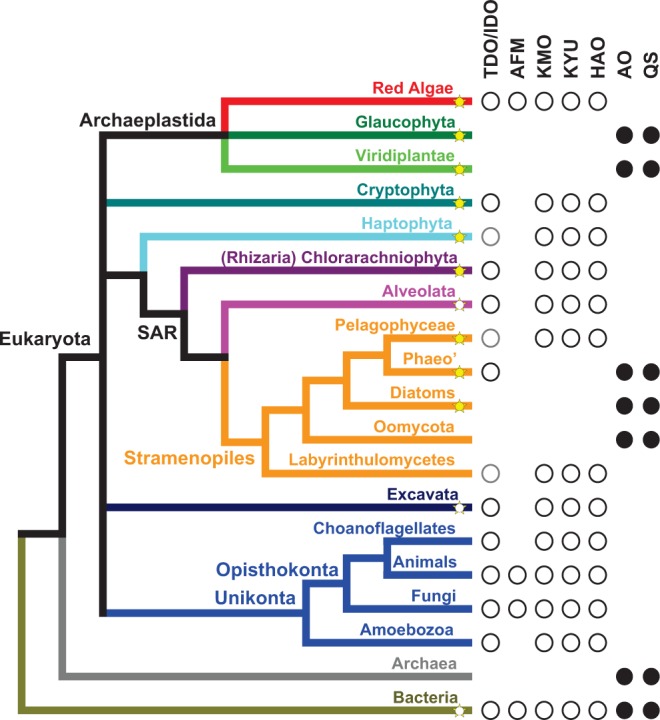
Phylogenetic distribution of de novo NAD+ biosynthesis pathways in eukaryotes shows a patchy distribution. Shown is the organismal phylogeny (Burki, Okamoto, et al. 2012; Keeling and Palmer 2008) for the major groups of life with emphasis on plastid bearing eukaryotes (star symbols; yellow, most species photoautotroph; white, some species photoautotroph; Phaeo’, Phaeophyceae or brown algae). For each clade, the presence of enzymes catalyzing the aspartate (closed symbols; AO, aspartate oxidase; QS, quinolinate synthase) or the kynurenine pathway (open symbols; TDO/IDO, tryptophan-/indoleamine 2,3-dioxygenase; AFM, arylformamidase; KMO, kynurenine 3-monooxygenase; KYU, kynureninase; HAO, 3-hydroxyanthranilate 3,4-dioxygenase) is indicated. Gray open symbols indicate that the IDO identified in this work, based on sequence similarity, might not actually catalyze the first reaction of the kynurenine pathway (see text). Most bacteria use the aspartate pathway, whereas some have the kynurenine pathway (see text). Very few bacteria, if any, seem to use both pathways in parallel.
