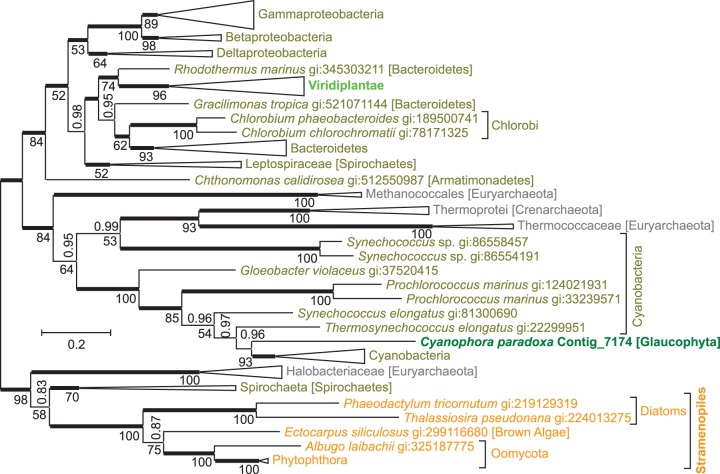
Fig. 3.—
An evolutionary tree for l-aspartate oxidase indicates separate origins of eukaryotic sequences. The unrooted Bayesian tree shows posterior probabilities above the branches and PhyML bootstrap values displayed as percentages below the branches. Thickened horizontal lines represent 1.0 Bayesian posterior probability. Trees with significantly lower log-likelihoods were obtained when monophyly was enforced for all sequences from Eukaryota, from any two of the three eukaryotic branches (Viridiplantae, Glaucophyta, Stramenopiles), or from Bacteria (supplementary table S1, Supplementary Material online). Larger clades have been collapsed for presentation and size is indicative of the number of taxa within the clade (full trees are available at TreeBase; http://purl.org/phylo/treebase/phylows/study/TB2:S15669, last accessed September 5, 2014). Scale bar represents 0.2 substitutions per site.