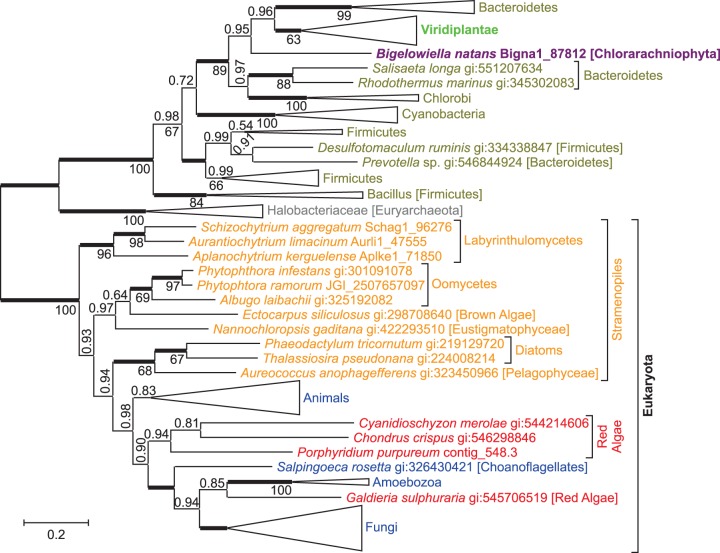
Fig. 5.—
An evolutionary tree for nicotinate-nucleotide pyrophosphorylase indicates that some photosynthetic eukaryotes acquired genes through transfers. The unrooted Bayesian tree shows posterior probabilities above the branches and PhyML bootstrap values below the branches. Thickened horizontal lines represent 1.0 Bayesian posterior probability. A tree with significantly lower log-likelihood was obtained when monophyly was enforced for all sequences from Eukaryota (supplementary table S1, Supplementary Material online). A tree with practically identical log-likelihood was obtained when monophyly was enforced for all sequences from Viridiplantae plus Bigelowiella natans (form a monophyletic clade within Bacteroidetes in PhyML tree). The sequence from Cyanophora paradoxa was omitted from an evolutionary analysis because it is incomplete. The best BLASTp hit for this incomplete sequence in the nr database from NCBI was with a nicotinate-nucleotide pyrophosphorylase sequence from Capsaspora owczarzaki (167 score, 67% coverage), and all top 100 BLASTp hits were with sequences from nonphotosynthetic eukaryotes, indicating possible descent of nicotinate-nucleotide pyrophosphorylase in Glaucophyta from the last common ancestor of eukaryotes.