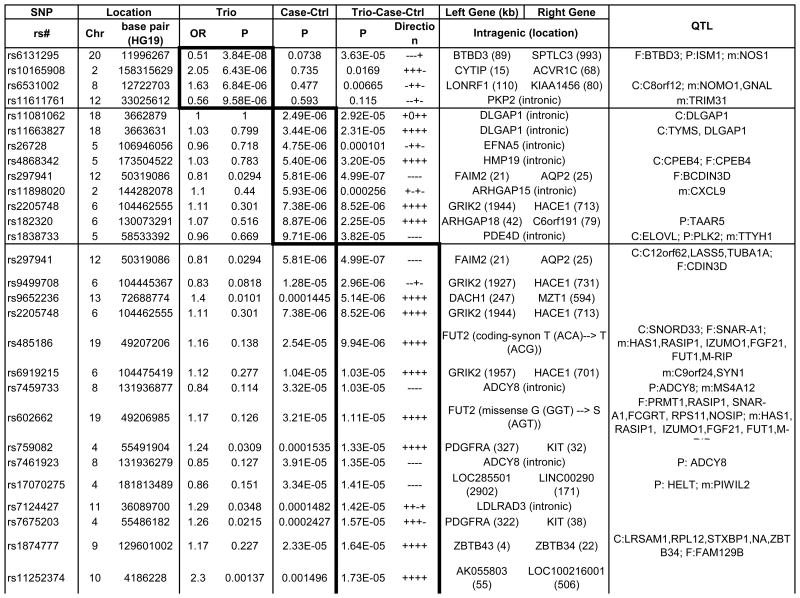
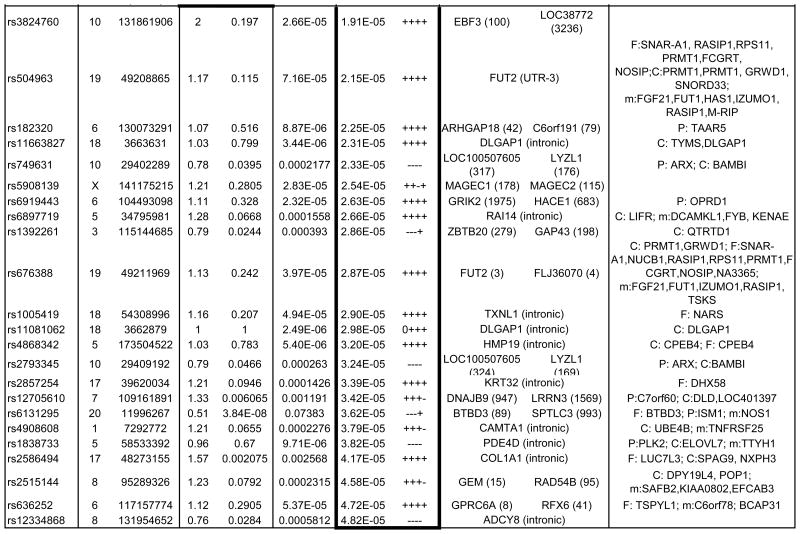
Table 1. Strongest Associated GWAS Variants in Trio, Case-Control and Combined Trio-Case-Control Samples.
Single nucleotide polymorphisms (SNP) listed by rs number include those with association P-values<10-5 for the trio and case-control samples, and those with P-values <5×10-5 for the combined trio-case-control sample association results. The chromosome (Chr) and base pair location for each SNP are listed in columns to the right of the SNP column. SNPs are listed separately for the analyses of trios (top section of table, with box around results), case-control samples including combined EU, SA and AJ MDS-defined ancestry subgroups (middle section and box), and for combined trio-case-control samples (lower section and box). SNPs with p<10-3 for any of the following are available in online Supplementary Table S2: EU, AJ and SA case-control subgroups individually and combined, trios, and combined case-control-trios). OR indicates the odds ratio for the tested allele in the trio sample. Direction indicates whether the direction of association between OCD and the A1 allele is either positive (+) or negative (-) for individual subgroups within the combined (EU, AJ, SA, trios) samples. The left gene and right gene columns lists the closest genes in the SNP region, either located within the gene (no distance given) or as right and left flanking genes (distance in kilobases). For SNPs located within genes, other functional elements in the region are as noted. QTL columns list genes whose expression (eQTL) or methylation levels (m) are associated (P-value) with the specified SNP in that row, specifically as identified previously in EU-ancestry frontal (F), parietal (P) or cerebellar (C) tissue. mQTL and F eQTL data were unavailable for X chromosome SNPs.
|
|
