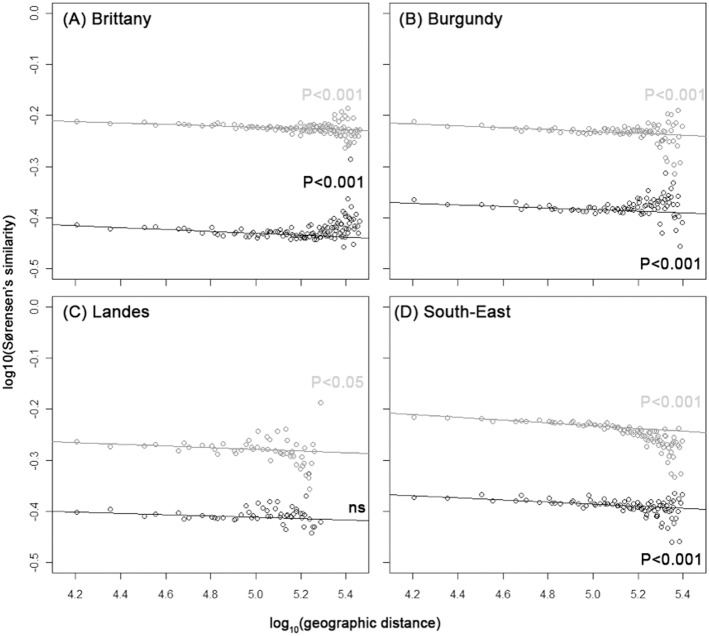
Figure 2. Distance-Decay Relationships for bacteria and fungi.
Each panel correspond to a region: Brittany (A), Burgundy (B), Landes (C) and South-East (D) Points the average Sørensen’s similarity between sites for each distance class. Lines represent the regression model based on the whole set of paired comparisons; for bacteria (grey) and fungi (black). The equations for the regression models were as follows: (A) Brittany: Bacteria***: log10(Sørensen’s similarity) = −0.014×log10(geographic distance)−0.156; Fungi***: log10(Sørensen’s similarity) = −0.017×log10(geographic distance)−0.350; (B) Burgundy: Bacteria*** log10(Sørensen’s similarity) = −0.018×log10(geographic distance)−0.144; Fungi***: log10(Sørensen’s similarity) = −0.015×log10(geographic distance)−0.316; (C) Landes: Bacteria*: log10(Sørensen’s similarity) = −0.017×log10(geographic distance)−0.198; Fungi ns: log10(Sørensen’s similarity) = −0.012×log10(geographic distance)−0.357; (D) South-East: Bacteria***: log10(Sørensen’s similarity) = −0.027×log10(geographic distance)−0.101; Fungi***: log10(Sørensen’s similarity) = −0.019×log10(geographic distance)−0.298. A graph with points representing all paired-comparisions between sites as points can be found in Figure S2. Significance of the model is indicated as an exponent for each organism: ns: not significant; P<0.05: *; P<0.01: **, P<0.001: ***.